Abstract
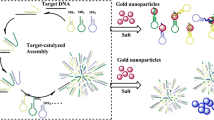
Taking advantage of recent developments in the field of metallic nanoparticle-based colorimetric DNA detection and in the field of in vitro selection of functional DNA/RNA that can recognize a wide range of analytes, we have designed highly sensitive and selective colorimetric biosensors for many analytes of choice. As an example of the sensor design strategy, a highly sensitive and selective colorimetric lead biosensor based on DNAzyme-directed assembly of gold nanoparticles is reviewed. The DNAzyme consists of an enzyme and a substrate strand, which can be used to assemble DNA-functionalized gold nanoparticles. The aggregation brings gold nanoparticles together, resulting in a blue-colored nanoparticle assembly. In the presence of lead, the DNAzyme catalyzes specific hydrolytic cleavage of the substrate strand, which disrupts the formation of the nanoparticle assembly, resulting in red-colored individual nanoparticles. The application of the sensor in lead detection in leaded paint is also demonstrated. In perspective, the use of allosteric DNA/RNAzymes to expand the range of the nanoparticle-based sensor design method is described.
Similar content being viewed by others
REFERENCES
H. B. Weiser (1933). Inorganic Colloid Chemistry, Vol. 1, Wiley, New York.
D. A. Handley (1989). In M. A. Hayat (Ed.), Coloidal Gold Principles, Methods, and Applications, Academic Press, San Diego, CA, pp. 1-12.
C. A. Mirkin, R. L. Letsinger, R. C. Mucic, and J. J. Storhoff (1996). A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 382(6592), 607-609.
R. Elghanian, J. J. Storhoff, R. C. Mucic, R. L. Letsinger, and C. A. Mirkin (1997). Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science 277(5329), 1078-1080.
J. J. Storhoff, R. Elghanian, R. C. Mucic, C. A. Mirkin, and R. L. Letsinger (1998). One-pot colorimetric differentiation of polynucleotides with single base imperfections using gold nanoparticle probes. J. Am. Chem. Soc. 120(9), 1959-1964.
T. A. Taton, C. A. Mirkin, and R. L. Letsinger (2000). Scanometric DNA array detection with nanoparticle probes. Science 289(5485), 1757-1760.
Y. Cao, R. Jin, and C. A. Mirkin (2001). DNA-modified core-shell Ag/Au nanoparticles. J. Am. Chem. Soc. 123(32), 7961-7962.
G. P. Mitchell, C. A. Mirkin, and R. L. Letsinger (1999). Programmed assembly of DNA functionalized quantum dots. J. Am. Chem. Soc. 121(35), 8122-8123.
R. Chakrabarti and A. M. Klibanov (2003). Nanocrystals modified with peptide nucleic acids (PNAs) for selective self-assembly and DNA detection. J. Am. Chem. Soc. 125(41), 12531-12540.
K. Sato, K. Hosokawa, and M. Maeda (2003). Rapid aggregation of gold nanoparticles induced by non-cross-linking DNA hybridization. J. Am. Chem. Soc. 125(27), 8102-8103.
L. Gold, B. Polisky, O. Uhlenbeck, and M. Yarus (1995). Diversity of oligonucleotide functions. Annu. Rev. Biochem. 64, 763-797.
S. E. Osborne and A. D. Ellington (1997). Nucleic acid selection and the challenge of combinatorial chemistry. Chem. Rev. 97(2), 349-370.
R. R. Breaker (1997). In vitro selection of catalytic polynucleotides. Chem. Rev. 97(2), 371-390.
D. S. Wilson and J. W. Szostak (1999). In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 68, 611-647.
G. F. Joyce and L. E. Orgel (1999). In R. F. Gesteland, T. R. Cech, and J. F. Atkins (Ed.), RNA World, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, pp. 49-77.
S. D. Jayasena (1999). Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 45(9), 1628-1650.
E. N. Brody and L. Gold (2000). Aptamers as therapeutic and diagnostic agents. Rev. Mol. Biotech. 74(1), 5-13.
J. Hesselberth, M. P. Robertson, S. Jhaveri, and A. D. Ellington (2000). In vitro selection of nucleic acids for diagnostic applications. Rev. Mol. Biotechnol. 74(1), 15-25.
G. A. Soukup and R. R. Breaker (2000). Allosteric nucleic acid catalysts. Curr. Opin. Struct. Biol. 10(3), 318-325.
R. R. Breaker (2002). Engineered allosteric ribozymes as biosensor components. Curr. Opin. Biotechnol. 13(1), 31-39.
H. Ueyama, M. Takagi, and S. Takenaka (2002). A novel potassium sensing in aqueous media with a synthetic oligonucleotide derivative. Fluorescence resonance energy transfer associated with guanine quartet-potassium ion complex formation. J. Am. Chem. Soc. 124(48), 14286-14287.
J. Ciesiolka and M. Yarus (1996). Small RNA-divalent domains. RNA 2(8), 785-793.
H. P. Hofmann, S. Limmer, V. Hornung, and M. Sprinzl (1997). Ni2+-binding RNA motifs with an asymmetric purine-rich internal loop and a G-A base pair. RNA 3(11), 1289-1300.
A. D. Ellington and J. W. Szostak (1992). Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 355(6363), 850-852.
A. D. Ellington and J. W. Szostak (1990). In vitro selection of RNA molecules that bind specific ligands. Nature 346(6287), 818-822.
C. Wilson and J. W. Szostak (1998). Isolation of a fluorophore-specific DNA aptamer with weak redox activity. Chem. Biol. 5(11), 609-617.
D. Grate and C. Wilson (1999). Laser-mediated, site-specific inactivation of RNA transcripts. Proc. Natl. Acad. Sci. U.S.A. 96(11), 6131-6136.
C. Wilson, J. Nix, and J. Szostak (1998). Functional requirements for specific ligand recognition by a biotin-binding RNA pseudoknot. Biochemistry 37(41), 14410-14419.
M. N. Stojanovic, P. de Prada, and D. W. Landry (2000). Fluorescent sensors based on aptamer self-assembly. J. Am. Chem. Soc. 122(46), 11547-11548.
G. R. Zimmermann, C. L. Wick, T. P. Shields, R. D. Jenison, and A. Pardi (2000). Molecular interactions and metal binding in the theophylline-binding core of an RNA aptamer. RNA 6(5), 659-667.
M. Meli, J. Vergne, J.-L. Decout, and M.-C. Maurel (2002). Adenine-aptamer complexes. A bipartite RNA site that binds the adenine nucleic base. J. Biol. Chem. 277(3), 2104-2111.
C. Mannironi, A. Di Nardo, P. Fruscoloni, and G. P. Tocchini-Valentini (1997). In vitro selection of dopamine RNA ligands. Biochemistry 36(32), 9726-9734.
I. Majerfeld and M. Yarus (1994). An RNA pocket for an aliphatic hydrophobe. Nat. Struct. Biol. 1(5), 287-292.
M. Famulok and J. W. Szostak (1992). Stereospecific recognition of tryptophan agarose by in vitro selected RNA. J. Am. Chem. Soc. 114(10), 3990-3991.
K. Harada and A. D. Frankel (1995). Identification of two novel arginine binding DNAs. EMBO J. 14(23), 5798-5811.
G. J. Connell, M. Illangesekare, and M. Yarus (1993). Three small ribooligonucleotides with specific arginine sites. Biochemistry 32(21), 5497-5502.
J. Tao and A. D. Frankel (1996). Arginine-binding RNAs resembling TAR identified by in vitro selection. Biochemistry 35(7), 2229-2238.
M. Famulok (1994). Molecular recognition of amino acids by RNA-aptamers: An L-citrulline binding RNA motif and its evolution into an L-arginine binder. J. Am. Chem. Soc. 116(5), 1698-1706.
G. J. Connell and M. Yarus (1994). RNAs with dual specificity and dual RNAs with similar specificity. Science 264(5162), 1137-1141.
N. K. Vaish, R. Larralde, A. W. Fraley, J. W. Szostak, and L. W. McLaughlin (2003). A novel, modification-dependent ATP-binding aptamer selected from an RNA library incorporating a cationic functionality. Biochemistry 42(29), 8842-8851.
M. Sassanfar and J. W. Szostak (1993). An RNA motif that binds ATP. Nature 364(6437), 550-553.
J. H. Davis and J. W. Szostak (2002). Isolation of high-affinity GTP aptamers from partially structured RNA libraries. Proc. Natl. Acad. Sci. U.S.A. 99(18), 11616-11621.
M. Koizumi and R. R. Breaker (2000). Molecular recognition of cAMP by an RNA aptamer. Biochemistry 39(30), 8983-8992.
S. M. Rink, J.-C. Shen, and L. A. Loeb (1998). Creation of RNA molecules that recognize the oxidative lesion 7,8-dihydro-8-hydroxy-2'-deoxyguanosine (8-oxodG) in DNA. Proc. Natl. Acad. Sci. U.S.A. 95(20), 11619-11624.
C. Boiziau, E. Dausse, L. Yurchenko, and J.-J. Toulme (1999). DNA aptamers selected against the HIV-1 trans-activation-responsive RNA element form RNA-DNA kissing complexes. J. Biol. Chem. 274(18), 12730-12737.
C. T. Lauhon and J. W. Szostak (1995). RNA aptamers that bind flavin and nicotinamide redox cofactors. J. Am. Chem. Soc. 117(4), 1246-1257.
P. Burgstaller and M. Famulok (1994). Isolation of RNA aptamers for biological cofactors by in vitro selection Angew. Chem. 106(10), 1163-1166 (see also Angew. Chem., Int. Ed. Engl. 1133(1110), 1084-1167(1994).
D. J. F. Chinnapen and D. Sen (2002). Hemin-stimulated docking of cytochrome c to a Hemin-DNA aptamer complex. Biochemistry 41(16), 5202-5212.
J. R. Lorsch and J. W. Szostak (1994). In vitro selection of RNA aptamers specific for cyanocobalamin. Biochemistry 33(4), 973-982.
M. Roychowdhury-Saha, S. M. Lato, E. D. Shank, and D. H. Burke (2002). Flavin recognition by an RNA aptamer targeted towardFAD. Biochemistry 41(8), 2492-2499.
D. Burke and D. Hoffman (1998). A novel acidophilic RNA motif that recognizes coenzyme A. Biochemistry 37(13), 4653-4663.
Y. Wang, J. Killian, K. Hamasaki, and R. R. Rando (1996). RNA molecules that specifically and stoichiometrically bind aminoglycoside antibiotics with high affinities. Biochemistry 35(38), 12338-12346.
M. G. Wallis, U. von Ahsen, R. Schroeder, and M. Famulok (1995). A novel RNA motif for neomycin recognition. Chem. Biol. 2(8), 543-552.
Q. Yang, I. J. Goldstein, H.-Y. Mei, and D. R. Engelke (1998). DNA ligands that bind tightly and selectively to cellobiose. Proc. Natl. Acad. Sci. U.S.A. 95(10), 5462-5467.
C. Srisawat, I. J. Goldstein, and D. R. Engelke (2001). Sephadex-binding RNA ligands: Rapid affinity purification of RNA from complex RNA mixtures. Nucleic Acids Res. 29(2), E4/1-E4/5.
S. T. Wallace and R. Schroeder (1998). In vitro selection and characterization of streptomycin-binding RNAs: Recognition discrimination between antibiotics. RNA 4(1), 112-123.
M. G. Wallis, B. Streicher, H. Wank, U. von Ahsen, E. Clodi, S. T. Wallace, M. Famulok, and R. Schroeder (1997). In vitro selection of a viomycin-binding RNA pseudoknot. Chem. Biol. 4(5), 357-366.
C. Berens, A. Thain, and R. Schroeder (2001). A tetracycline-binding RNA aptamer. Bioorg. Med. Chem. 9(10), 2549-2556.
L. Giver, D. Bartel, M. Zapp, A. Pawul, M. Green, and A. D. Ellington (1993). Selective optimization of the Rev-binding element of HIV-1. Nucleic Acids Res. 21(23), 5509-5516.
K. P. Williams, X.-H. Liu, T. N. M. Schumacher, H. Y. Lin, D. A. Ausiello, P. S. Kim, and D. P. Bartel (1997). Bioactive and nuclease-resistant L-DNA ligand of vasopressin. Proc. Natl. Acad. Sci. U.S.A. 94(21), 11285-11290.
D. Nieuwlandt, M. Wecker, and L. Gold (1995). In vitro selection of RNA ligands to substance P. Biochemistry 34(16), 5651-5659.
L. C. Bock, L. C. Griffin, J. A. Latham, E. H. Vermaas, and J. J. Toole (1992). Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 355(6360), 564-566.
C. Tuerk, S. MacDougal, and L. Gold (1992). RNA pseudoknots that inhibit human immunodeficiency virus type 1 reverse transcriptase. Proc. Natl. Acad. Sci. U.S.A. 89(15), 6988-6992.
M. Vuyisich and P. A. Beal (2002). Controlling protein activity with ligand-regulated RNA aptamers. Chem. Biol. 9(8), 907-913.
F. Pileur, M.-L. Andreola, E. Dausse, J. Michel, S. Moreau, H. Yamada, S. A. Gaidamakov, R. J. Crouch, J.-J. Toulme, and C. Cazenave (2003). Selective inhibitory DNA aptamers of the human RNase H1. Nucleic Acids Res. 31(19), 5776-5788.
N. C. Pagratis, C. Bell, Y.-F. Chang, S. Jennings, T. Fitzwater, D. Jellinek, and C. Dang (1997). Potent 2'-amino-, and 2'-fluoro-2'-deoxyribonucleotide RNA inhibitors of keratinocyte growth factor. Nature Biotech. 15(1), 68-73.
D. Jellinek, L. S. Green, C. Bell, C. K. Lynott, N. Gill, C. Vargeese, G. Kirschenheuter, D. P. C. McGee, P. Abesinghe, et al. (1995). Potent 2'-amino-2'-deoxypyrimidine RNA inhibitors of basic fibroblast growth factor. Biochemistry 34(36), 11363-11372.
J. Ruckman, L. S. Green, J. Beeson, S. Waugh, W. L. Gillette, D. D. Henninger, L. Claesson-Welsh, and N. Janjic (1998). 2'-fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 273(32), 20556-20567.
L. L. Lebruska and L. J. MaherIII (1999). Selection and characterization of an RNA decoy for transcription factor NF-ΚB. Biochemistry 38(10), 3168-3174.
T. W. Wiegand, P. B. Williams, S. C. Dreskin, M. H. Jouvin, J. P. Kinet, and D. Tasset (1996). High-affinity oligonucleotide ligands to human IgE inhibit binding to Fc epsilon receptor I. J. Immun. 157(1), 221-230.
I. A. Nazarenko and O. C. Uhlenbeck (1995). Defining a smaller RNA substrate for elongation factor Tu. Biochemistry 34(8), 2545-2552.
K. A. Davis, Y. Lin, B. Abrams, and S. D. Jayasena (1998). Staining of cell surface human CD4 with 2'-F-pyrimidine-containing RNA aptamers for flow cytometry. Nucleic Acids Res. 26(17), 3915-3924.
S. Jeong, T.-Y. Eom, S.-J. Kim, S.-W. Lee, and J. Yu (2001). In vitro selection of the RNA aptamer against the Sialyl Lewis X and its inhibition of the cell adhesion. Biochem. Biophys. Res. Commun. 281(1), 237-243.
M. Blank, T. Weinschenk, M. Priemer, and H. Schluesener (2001). Systematic evolution of a DNA aptamer binding to rat brain tumor microvessels. Selective targeting of endothelial regulatory protein pigpen. J. Biol. Chem. 276(19), 16464-16468.
W. Pan, R. C. Craven, Q. Qiu, C. B. Wilson, J. W. Wills, S. Golovine, and J.-F. Wang (1995). Isolation of virus-neutralizing RNAs from a large pool of random sequences. Proc. Natl. Acad. Sci. U.S.A. 92(25), 11509-11513.
J. G. Bruno and J. L. Kiel (1999). In vitro selection of DNA aptamers to anthrax spores with electrochemiluminescence detection. Biosens. Bioelectr. 14(5), 457-464.
J. Tsang and G. F. Joyce (1996). In vitro evolution of randomized ribozymes. Methods Enzymol. 267 (Combinatorial Chemistry), 410-426.
R. R. Breaker (1997). DNA enzymes. Nat. Biotechnol. 15(5), 427-431.
R. R. Breaker (1997). DNA aptamers and DNA enzymes. Curr. Opin. Chem. Biol. 1(1), 26-31.
D. Sen and C. R. Geyer (1998). DNA enzymes. Curr. Opin. Chem. Biol. 2(6), 680-687.
M. Kurz and R. R. Breaker (1999). In vitro selection of nucleic acid enzymes. Current Topics in Microbiology and Immunology 243 (Combinatorial Chemistry in Biology), 137-158.
T. R. Cech (1987). The chemistry of self-splicing RNA and RNA enzymes. Science 236(4808), 1532-1539.
T. R. Cech (2000). Perspectives. Structural biology: The ribosome is a ribozyme. Science 289(5481), 878-879.
N. K. Vaish, P. A. Heaton, O. Fedorova, and F. Eckstein (1998). In vitro selection of a purine nucleotide-specific hammerhead-like ribozyme. Proc. Natl. Acad. Sci. U.S.A. 95(5), 2158-2162.
E. H. Ekland, J. W. Szostak, and D. P. Bartel (1995). Structurally complex and highly active RNA ligases derived from random RNA sequences. Science 269(5222), 364-370.
J. R. Lorsch and J. W. Szostak (1994). In vitro evolution of new ribozymes with polynucleotide kinase activity. Nature 371(6492), 31-36.
E. H. Ekland and D. P. Bartel (1996). RNA-catalyzed RNA polymerization using nucleoside triphosphates. Nature 382(6589), 373-376.
M. Illangasekare and M. Yarus (1997). Small-molecule-substrate interactions with a self-aminoacylating ribozyme. J. Mol. Biol. 268(3), 631-639.
J. A. Piccirilli, T. S. McConnell, A. J. Zaug, H. F. Noller, and T. R. Cech (1992). Aminoacyl esterase activity of the Tetrahymena ribozyme. Science 256(5062), 1420-1424.
P. A. Lohse and J. W. Szostak (1996). Ribozyme-catalyzed amino-acid transfer reactions. Nature 381(6581), 442-444.
C. Wilson and J. W. Szostak (1995). In vitro evolution of a self-alkylating ribozyme. Nature 374(6525), 777-782.
M. Wecker, D. Smith, and L. Gold (1996). In vitro selection of a novel catalytic RNA: Characterization of a sulfur alkylation reaction and interaction with a small peptide. RNA 2(10), 982-994.
X. Dai, A. De Mesmaeker, and G. F. Joyce (1995). Cleavage of an amide bond by a ribozyme. Science 267(5195), 237-240.
T. W. Wiegand, R. C. Janssen, and B. E. Eaton (1997). Selection of RNA amide synthases. Chem. Biol. 4(9), 675-683.
B. Zhang and T. R. Cech (1997). Peptide bond formation by in vitro selected ribozymes. Nature 390(6655), 96-100.
T. M. Tarasow, S. L. Tarasow, C. Tu, E. Kellogg, and B. E. Eaton (1999). Characteristics of an RNA diels-alderase active site. J. Am. Chem. Soc. 121(15), 3614-3617.
J. R. Prudent, T. Uno, and P. G. Schultz (1994). Expanding the scope of RNA catalysis. Science 264(5167), 1924-1927.
M. M. Conn, J. R. Prudent, and P. G. Schultz (1996). Porphyrin metalation catalyzed by a small RNA molecule. J. Am. Chem. Soc. 118(29), 7012-7013.
R. R. Breaker and G. F. Joyce (1994). A DNA enzyme that cleaves RNA. Chem. Biol. 1(4), 223-229.
R. R. Breaker (2000). Making catalytic DNAs. Science 290(5499), 2095-2096.
Y. Lu (2002). New transition metal-dependent DNAzymes as efficient endonucleases and as selective metal biosensors. Chem. Eur. J. 84588-4596.
R. R. Breaker and G. F. Joyce (1995). A DNA enzyme with Mg2+-dependent RNA phosphoesterase activity. Chem. Biol. 2(10), 655-660.
D. Faulhammer and M. Famulok (1997). Characterization and divalent metal-ion dependence of in vitro selected deoxyribozymes which cleave DNA/RNA chimeric oligonucleotides. J. Mol. Biol. 269(2), 188-202.
S. W. Santoro and G. F. Joyce (1997). A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. U. S. A. 94(9), 4262-4266.
C. R. Geyer and D. Sen (1997). Evidence for the metal-cofactor independence of an RNA phosphodiester-cleaving DNA enzyme. Chem. Biol. 4(8), 579-593.
A. Roth and R. R. Breaker (1998). An amino acid as a cofactor for a catalytic polynucleotide. Proc. Natl. Acad. Sci. U.S.A. 95(11), 6027-6031.
J. Li, W. Zheng, A. H. Kwon, and Y. Lu (2000). In vitro selection and characterization of a highly efficient Zn(II)-dependent RNA-cleaving deoxyribozyme. Nucleic Acids Res. 28(2), 481-488.
N. Carmi, L. A. Shultz, and R. R. Breaker (1996). In vitro selection of self-cleaving DNAs. Chem. Biol. 3(12), 1039-1046.
B. Cuenoud and J. W. Szostak (1995). A DNA metalloenzyme with DNA ligase activity. Nature 375(6532), 611-614.
Y. Wang and S. K. Silverman (2003). Deoxyribozymes that synthesize branched and lariat RNA. J. Am. Chem. Soc. 125(23), 6880-6881.
Y. Li and R. R. Breaker (1999). Phosphorylating DNA with DNA. Proc. Natl. Acad. Sci. U.S.A. 96(6), 2746-2751.
Y. Li, Y. Liu, and R. R. Breaker (2000). Capping DNA with DNA. Biochemistry 39(11), 3106-3114.
Y. Li and D. Sen (1996). A catalytic DNA for porphyrin metallation. Nat. Struct. Biol. 3(9), 743-747.
D. Faulhammer and M. Famulok (1996). The Ca2+ ion as a cofactor for a novel RNA-cleaving deoxyribozyme. Angew. Chem., Int. Ed. Engl. 35(23/24), 2837-2841.
J. J. Storhoff, A. A. Lazarides, R. C. Mucic, C. A. Mirkin, R. L. Letsinger, and G. C. Schatz (2000). What controls the optical properties of DNA-linked gold nanoparticle assemblies? J. Am. Chem. Soc. 122(19), 4640-4650.
R. Jin, G. Wu, Z. Li, C. A. Mirkin, and G. C. Schatz (2003). What controls the melting properties of DNA-linked gold nanoparticle assemblies? J. Am. Chem. Soc. 125(6), 1643-1654.
J. Liu and Y. Lu (2003). A colorimetric lead biosensor using DNAzyme-directed assembly of gold nanoparticles. J. Am. Chem. Soc. 125(22), 6642-6643.
L. M. Demers, C. A. Mirkin, R. C. Mucic, R. A. Reynolds, III, R. L. Letsinger, R. Elghanian, and G. Viswanadham (2000). A fluorescence-based method for determining the surface coverage and hybridization efficiency of thiol-capped oligonucleotides bound to gold thin films and nanoparticles. Anal. Chem. 72(22), 5535-5541.
J. Liu and Y. Lu (2003). Improving fluorescent DNAzyme biosensors by combining inter-and intramolecular quenchers. Anal. Chem. 75(23), 6666-6672.
U. S. Department of Housing and Urban Development (2001). Unpublished data, 2001.
K. K. Luk, L. L. Hodson, J. A. O'Rouke, D. S. Smith, and W. F. Gutknecht (1993). Investigation of Test Kits for Detection of Lead in Paint, Dust, and Soil, EPA 600/R-93/085, U.S. Environmental Protection Agency, Research Triangle Park, NC.
W. J. Rossiter,Jr., M. G. Vangel, M. E. McKnight, and G. Dewalt (2000, March). Spot Test Kits for Detecting Lead in Household Paint: A Laboratory Evaluation, NISTIR 6398, National Institute of Standards and Technology, Gaithersburg, MD.
A. W. Czarnik (1995). Desperately seeking sensors. Chem. Biol. 2(7), 423-428.
J. Tang and R. R. Breaker (1997). Rational design of allosteric ribozymes. Chem. Biol. 4(6), 453-459.
D. Y. Wang, B. H. Y. Lai, and D. Sen (2002). A general strategy for effector-mediated control of RNA-cleaving ribozymes and DNA enzymes. J. Mol. Biol. 318(1), 33-43.
M. Levy and A. D. Ellington (2002). ATP-dependent allosteric DNA enzymes. Chem. Biol. 9(4), 417-426.
G. A. Soukup, G. A. M. Emilsson, and R. R. Breaker (2000). Altering molecular recognition of RNA aptamers by allosteric selection. J. Mol. Biol. 298(4), 623-632.
G. A. Soukup, E. C. DeRose, M. Koizumi, and R. R. Breaker (2001). Generating new ligand-binding RNAs by affinity maturation and disintegration of allosteric ribozymes. RNA 7(4), 524-536.
D. E. Huizenga and J. W. Szostak (1995). A DNA aptamer that binds adenosine and ATP. Biochemistry 34(2), 656-665.
J. Liu and Y. Lu (2004). Adenosine dependent assembly of aptazyme-functionalized gold nanoparticles and their application as a colorimetric biosensor. Anal. Chem. 76(6), 1627-1632.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, J., Lu, Y. Colorimetric Biosensors Based on DNAzyme-Assembled Gold Nanoparticles. Journal of Fluorescence 14, 343–354 (2004). https://doi.org/10.1023/B:JOFL.0000031816.06134.d3
Issue Date:
DOI: https://doi.org/10.1023/B:JOFL.0000031816.06134.d3