Abstract
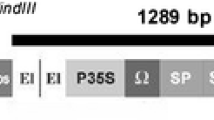
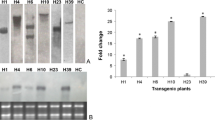
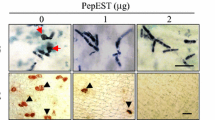
Phytophthora citrophthora is the most widely spread oomycete plant pathogen over all the citrus growing areas and represents one of the major causes of crop losses. Constitutive over-expression of genes encoding proteins involved in plant defence mechanisms to disease is one of the strategies proposed to increase plant tolerance to oomycete and fungal pathogens. P23 (PR-5), a 23-kDa pathogenesis-related protein similar to osmotins, is induced in tomato (Lycopersicon esculentum Mill. cv. Rutgers) plants when they are infected with citrus exocortis viroid, and its antifungal activity has been demonstrated in in vitro assays. We have successfully produced transgenic orange (Citrus sinensis L. Obs. cv. Pineapple) plants bearing a chimeric gene construct consisting of the cauliflower mosaic virus 35S promoter and the coding region of the tomato pathogenesis-related PR-5. Nine regenerated transgenic lines constitutively expressed the PR protein. They were challenged with Phytophthora citrophthora using a detached bark assay. A significant reduction in lesion development was consistently observed in one transgenic line in comparison to the control plants. This same line achieved plant survival rates higher than control plants when transgenic trees were inoculated with oomycete cultures. These results provide evidence for the in vivo activity of the tomato PR-5 protein against Phytophthora citrophthora, and suggest that this may be employed as a strategy aimed at engineering Phytophthora disease resistance in citrus.
Similar content being viewed by others
References
Alexander D, Goodman RM, Gut-Rella M, Glascock C, Weymann K, Friedrich L, Maddox D, Ahl-Goy P, Luntz T, Ward E, Ryals J: Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1 a. Proc Natl Acad Sci USA 90: 7327–7331 (1993).
Bradford MM: A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 (1976).
Broglie K, Chet Y, Holliday M, Cressman R, Biddle PH, Knowlton S, Mauvais J, Broglie R: Transgenic plants with enhanced resistance to the fungal pathogen Rhizotocnia solani. Science 254: 1194–1197 (1991).
Dellaporta SL, Wood J, Hicks JB: A plant DNA minipreparation: Version II. Plant Mol Biol Rep 4: 19–21 (1983).
Erwin DC, Ribeiro OK: Phytophthora diseases worlwide. American Phytopathological Society, St. Paul, MN (1996).
Evans IJ, Greenland A: Trangenic approaches to disease protection: Applications of antifungal proteins. Pestic Sci 54: 353–359 (1998).
Granell A, Belles JM, Conejero V: Induction of pathogenesisrelated proteins in tomato by citrus exocortis viroid, silver ions and ethephon. Physiol Mol Plant Pathol 31: 83–89 (1987).
Grison R, Grezes-Besset B, Scheneider M, Lucante N, Olsen L, Leguay JJ, Toppan A: Field tolerance to fungal pathogens of Brassica napus constitutively expressing a chimeric chitinase gene. Nature Biotechnology 14: 643–646 (1996).
Heller WE, Gessler C: Induced systemic resistance in tomato plants against Phytophthora infestans. J Phytopathol 116: 323–328 (1986).
Hood EE, Gelvin SB, Melchers LS, Hoekema A: New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res 2: 208–218 (1993).
Jefferson RA: Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5: 387–405 (1987).
Jongedijk E, Tigelaar H, van Roekel JSC, Bres-Vloemans SA, Dekker I, van den Elzen PJM, Cornelissen BJC, Melchers LS: Synergistic activity of chitinases and β-1,3-glucanases enhances fungal resistance in transgenic tomato plants. Euphytica 85: 173–180 (1995).
Judelson HS: Recent advances in the Genetics of Oomycete plant pathogens. Mol Plant Microb Interac 9: 443–449 (1996).
Kamoun S, Huitema E, Vleeshouwers VGAA: Resistance to oomycetes: a general role for the hypersensitive response?. Trends in Plant Sci 4: 196–200 (1999).
Lin W, Anuratha CS, Datta K, Potrykus I, Muthukrishnan S, Datta SK: Genetic engineering of rice for resistance to Sneath blight. Bio/technology 13: 686–691 (1995).
Liu D, Raghothama KG, Hasegawa PM, Bressan RA: Osmotin overexpression in potato delays development of disease symptoms. Proc Natl Acad Sci USA 91: 1888–1892 (1994).
Mauch F, Mauch-Mani B, Boller T: Antifungal hydrolases in pea tissue. Plant Physiol 88: 936–942 (1988).
McIntyre JL, Dodds JA: Induction of localized and systemic protection against Phytophthora parasitica var. nicotianae by tobacco mosaic virus infection of tobacco hypersensitive to the virus. Physiol Plant Pathol 15: 321–330 (1979).
Murashige T, Skoog F: A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiol Plant 15: 473–479 (1962).
Peña L, Cervera M, Juárez J, Navarro A, Pina JA, Durán-Vila N, Navarro L: Agrobacterium-mediated transformation of sweet orange and regeneration of transgenic plants. Plant Cell Rep 14: 616–619 (1995).
Peña L, Navarro L: Transgenic citrus. In: Bajaj YPS (ed.) Transgenic Trees, pp. 39–54. Springer-Verlag, Berlin (1999).
Punja ZK, Raharjo HT: Response of transgenic cucumber and carrot plants expressing different chitinase enzymes to inoculation with fungal pathogens. Plant Dis 80: 999–1005 (1996).
Roberts WK, Selitrennikoff CP: Zeamatin, an antifungal protein from maize with membrane-permeabilizing activity. J Gen Microbiol 136: 1771–1778 (1990).
Rodrigo I, Vera P, Frank R, Conejero, V: Identification of the viroid-induced tomato pathogenesis-related (PR) protein P23 as the thaumatin-like tomato protein NP24 associated with osmotic stress. Plant Mol Biol 16: 931–934 (1991).
Rodrigo I, Vera P, Tornero P, Hernandez-Yago J, Conejero, V: cDNA cloning of viroid-induced tomato pathogenesis-related protein P23. Characterization as a vacuolar antifungal factor. Plant Physiol 102: 939–945 (1993).
Sambrook J, Fritsch EF, Maniatis T: Molecular cloning - a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor (1989).
SAS Institute: SAS/STAT User's Guide, release 6.03. SAS Institute, Cary, NC (1991).
Schlumbaum A, Mauch F, Vogëli U, Boller T: Plant chitinases are potent inhibitors of fungal growth. Nature 324: 365–367 (1986).
Tabaeizadeh Z, Agharbaoui Z, Harrak H, Poysa V: Transgenic tomato plants expressing a Lycopersicon chilense chitinase gene demonstrate improved resistance to Verticillium dahliae race 2. Plant Cell Rep 19: 197–202 (1999).
Terakawa T, Takaya N, Horiuchi H, Koike M, Takagi M: A fungal chitinase gene from Rhizopus oligosporus confers antifungal activity to transgenic tobacco. Plant Cell Rep 16: 439–443 (1997).
Timmer LW, Menger JA: Phytophthora-induced diseases. In: Whiteside JO, Garnsey SM, Timmer LW (eds) Compendium of Citrus Diseases, pp. 22–24. Am Phytopathol Soc Press, St. Paul, MN (1988).
Tuset JJ, Hinarejos C, Garcia J: Present status of Phytophthora diseases of citrus in Spain. Proc Int Soc Citriculture 1: 338–343 (1984).
Van Loon LC, Pierpoint WS, Boller T, Conejero V: Recommendations for naming plant pathogenesis-related proteins. Plant Mol Biol Rep 12: 245–264 (1994).
Van Loon LC, Van Strien EA: The families of pathogenesisrelated proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol 55: 85–97 (1999).
Woloshuk CP, Meulenhoff JS, Sela-Buurlage M, van den Elzen PJM, Cornelissen BJC: Pathogen induced proteins with inhibitory activity toward Phytophthora infestans. Plant Cell 3: 619–628 (1991).
Zhu B, Chen THH, Li PH: Analysis of late-blight disease resistance and freezing tolerance in transgenic potato plants expressing sense and antisense genes for an osmotin-like protein. Planta 198: 70–77 (1996).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Fagoaga, C., Rodrigo, I., Conejero, V. et al. Increased tolerance to Phytophthora citrophthora in transgenic orange plants constitutively expressing a tomato pathogenesis related protein PR-5. Molecular Breeding 7, 175–185 (2001). https://doi.org/10.1023/A:1011358005054
Issue Date:
DOI: https://doi.org/10.1023/A:1011358005054