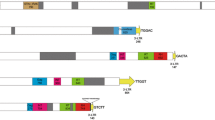
Abstract
A 1 kb EcoRI restriction fragment cloned from a band visible in an agarose gel of Pinus lambertiana (sugar pine) genomic DNA is present in both subgenera of Pinus with at least 104 copies/genome. A full-length copy of this repeated element recovered from a P. radiata (Monterey pine) genomic DNA library was found to possess all of the sequence features associated with gypsy-like retrotransposons. This report describes the biology and history of the IFG (Institute of Forest Genetics) family of retrotransposons. The characterized IFG7 is 5937 bp long. Immediately interior to its 5′ and 3′ long terminal repeats are sequences consistent with primer binding sites for reverse transcription of the RNA genome. Presumptive gene products associated with retrotransposition appear to be coded in a single reading frame and are in the same order as the gypsy-like retrotransposons and retroviruses. The 1.0 kb EcoRI fragment of IFG elements codes for the 3′ half of IFG's reverse transcriptase and the entire RNase H domain. Southern blot analysis suggests IFG was present in Pinaceae before its division into its modern genera. Sequence analysis of IFG 1.0 kb RI fragments and southern analysis also suggest that IFG continued to evolve in Pinus with restriction fragment length polymorphism (RFLP) subfamilies appearing early in the history of each subgenus often correlating with subdivisions of Pinus. Features shared with other plant retrotransposons are also discussed.
Similar content being viewed by others
References
Axelrod DI: Cenozoic history of some western American pines. Ann Miss Bot Gard 73: 565–641(1986).
Berg JM: Potential metal-binding domains in nucleic acid binding proteins. Science232: 485–487(1986).
Bingham PM, Zachar Z: Retrotransposons and the FB Trans-poson from Drosophila melanogaster. In: Berg DE, Howe MM (eds) Mobile DNA, pp. 450–503, American Society for Microbiology, Washington, D.C. (1989).
Boeke JD: Transposable elements in Saccharomyces cere-visiae In: Berg DE, Howe MM (eds) Mobile DNA, pp. 335–374, American Society for Microbiology, Washington, D.C. (1989).
Camirand A and Brisson N: The complete nucleotide sequence of the Tst1 retrotransposon of potato. Nucl Acids Res 18: 4929 (1990).
Chiu LM, Callahan R, Tronick SR, Schlom J, Aaronson S.A: Major pol gene progenitors in the evolution of oncoviruses. Science 223: 364–370(1984).
Donehower L, Gillespie D: Restriction site periodicities in highly repetitive DNA of primates. J Mol Biol 134: 805–834 (1979).
Doolittle RF, Feng DF, Johnson MJ, McClure MA: Origins and evolutionary relationships of retroviruses. Q Rev Biol 64: 1–30(1989).
Feinberg AO, Vogelstein B: A technique for radiolabeling DNA restriction endonuclease fragments to high specific ac-tivity. Anal Biochem 132: 6–13(1983).
Friesen PD, Nissen MS: Gene organization and transcription of TED, a Lepidopteran retrotransposon integrated within the baculovirus genome. Mol Cell Biol 10: 3067–3077(1990).
Grandbastien M, Spielmann A, Caboche M: Tnt1, a mobile retroviral-like transposable element of tobacco isolated by plant cell genetics. Nature 337: 376–380(1989).
Hansen LJ, Chalker DL, Sandmeyer SB: Ty3, a yeast retro-transposon associated with tRNA genes, has homology to animal retroviruses. Mol Cell Biol 8: 5245–5256(1988).
Henikoff S: Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene 28: 351–359(1984).
Jin YK, Bennetzen JL: Structure and coding properties of Bs1, a maize retrovirus-like transposon. Proc Natl Acad Sci USA 86: 6235–6239(1989).
Johnson MS, McClure MA, Feng DF, Gray J, Doolittle RF: Computer analysis of retrovirus pol genes: assignment of en-zymatic function to specific sequences and homologies with nonviral enzymes. Proc Natl Acad Sci USA 83: 7648–7652 (1986).
Kamm A, Doudrick RL, Heslop-Harrison JS, Schmidt T: The genomic and physical organization of Ty1-copia-like se-quences as a component of large genomes in Pinus elliottii var. elliottii and other gymnosperms. Proc Natl Acad Sci USA 93: 2708–2713(1996).
Kriebel HB DNA sequence components of the Pinus strobus nuclear genome. Can J For Res 15: 1–4(1985).
Langley CH., Montgomery E, Hudson R, Kaplan N, Charlesworth B: On the role of unequal exchange in the con-tainment of transposable element copy number. Genet Res 52: 223–235(1988).
Li WH., Wu CI, Luo CC: A new method for estimating syn-onymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol. Biol. Evol. 2: 150–174(1985).
Lindauer A, Fraser D, Brüderlein M, Schmitt R: Reverse tran-scriptase families and a copia-like retrotransposon, Osser, in the green alga Volvox carteri. FEBS J 319: 261–266(1993).
Little EL, Critchfield WB: Subdivisions of the Genus Pi-nus (pines). U.S. Department of Agriculture Miscellaneous Publications 1144 (1969).
Maniatis T, Fritsch EF, Sambrook J: Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY (1982).
Marlor RL, Parkhurst SM, Corces VG: The Drosophila melanogaster gypsy transposable element encodes putative gene products homologous to retroviral proteins. Mol Cell Biol 6: 1129–1134(1986).
McAllister BF, Werren JH: Phylogenetic analysis of a retro-transposon with implications for strong evolutionary con-straints on reverse transcriptase. Mol Biol Evol 14: 69–80 (1997).
Mirov NT: The Genus Pinus. Ronald, New York (1967).
Mount S, Rubin G: Complete nucleotide sequence of the Drosophila transposable element copia: homology between copia and retroviral proteins. Mol Cell Biol5: 1630–1638 (1985).
Patarca R, Haseltine WA: Sequence similarity among retro-viruses. Nature 309: 728 (1984).
Pearl LH, Taylor WR: A structural model for the retroviral proteases. Nature 329: 351- 354 (1987).
Prats AC, Sarih L, Gabus C, Litvak S, Keith G, Darlix JL: Small finger protein of avian and murine retroviruses has nucleic acid annealing activity and positions the replication primer tRNA onto genomic RNA. EMBO J 7: 1777–1783 (1988).
Price RA, Olsen-Stojkovich J, Lowenstein JM: Relationships among the genera of Pinaceae: an immunological comparison. Syst Bot 12: 91–97(1987).
Roeder GS, Fink GR: Transposable elements in yeast. In: Shapiro J (ed) Mobile Genetic Elements, pp. 299–326. Aca-demic Press, New York (1983).
Rogers SO, Bendich AJ: Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol Biol 5: 69–76(1985).
Sederoff, R., Stomp A-M, Gwynn G, Ford E, Loopstra C, Hodgskiss P, Chilton WS: Application of recombinant DNA techniques to pines: A molecular approach to genetic engi-neering in forestry. In: Bonga JM, Durzan DJ (eds) Cell and Tissue Culture in Forestry, pp. 314–329. Kluwer Academic Publishers, Dordrecht, Netherlands (1987).
Shinnick T, Lerner R, Sutcliffe J. Nucleotide sequence of Moloney murine leukemia virus. Nature 293: 543–548(1981).
Smyth DR, Kalitsis P, Joseph JL, Sentry JW: Plant retrotrans-poson from Lilium henryi is related to Ty3 of yeast and the gypsy group of Drosophila. Proc Natl Acad Sci USA, 86: 5015–5019(1989).
Sprinzl M, Hartman T, Weber J, Blank J, Zeidler R: Compila-tion of tRNA sequences and sequences of tRNA genes. Nucl Acids Res 17 (Suppl) 1–72(1989).
Strauss SH, Doerksen AH: Restriction fragment analysis of pine phylogeny. Evolution 44: 1081–1096(1990).
Su PY, Brown TA: Ty3/gypsy-like retrotransposon sequences in tomato. Plasmid 38: 148–157(1997).
Swofford DL: PAUP: Phylogenetic analysis using parsimony, Version 3.0L (Mac). Illinois Natural History Survey, Cham-paign, IL (1990).
Van der Burgh J: Woods of the Rhenish brown-coal formation, 2. Woods, from the brown-coal pits 'Maria Theresia' at Herzo-genrath, 'Zukunft West' at Eschweiler and 'Victor' ( Zülpich Mitte) at Zülpich. With a systematic-anatomical study of the genus Pinus L. Rev Paleobot Palynol 15: 73–275(1973).
Varmus H, Brown P: Retroviruses. In: Berg DE, Howe MM (eds) Mobile DNA, pp. 53–108. American Society for Micro-biology, Washington, D.C. (1989).
Voytas DF, Ausubel FM: A copia-like transposable element family in Arabidopsis thaliana. Nature 336: 242–244(1988).
Voytas DF, Cummings MP, Konieczny A, Ausubel FM, Ro-dermel SR: Copia-like retrotransposons are ubiquitous among plants. Proc Natl Acad Sci USA 89: 7124–7128(1992).
Voytas DF, Konieczny A, Commings MP, Ausubel FM: The structure, distribution and evolution of the Ta1 retrotranspos-able element family of Arabidopsis thaliana. Genetics 126: 713–721(1990).
Wakainiya I, Newton RJ, Johnston JS, Price, HJ: Genome size and environmental factors in the genus Pinus. Am J Bot 80:1235–1241(1993).
Wolfe KH, Sharp PM, Li W-H: Rates of synonymous sub-stitution in plant nuclear genes. J Mol Evol 29: 208- 211 (1989).
Xiong Y, Eickbush T: Origin and evolution of retroelements based upon their reverse transcriptase sequence. EMBO 9: 3353–3362(1990).
Yuki S, Ishimaru S, Inouye S, Saigo K: Identification of genes for reverse transcriptase-like enzymes in two Drosophila retrotransposons 412 and gypsy; a rapid detection method of reverse transcriptase genes using YXDD box probes. Nucl Acids Res. 14: 3017–3030(1986b).
Zararin E: Taxonomy of pinyons based on terpenoids from wood oleoresin and on morphological and other biological characters. In: Proceedings Second International Pinyon Pine Symposium (August 6- 8, 1987), Lomas de Chapultepec, Mexico, D.F. (1989).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Kossack, D.S., Kinlaw, C.S. IFG, a gypsy-like retrotransposon in Pinus (Pinaceae), has an extensive history in pines. Plant Mol Biol 39, 417–426 (1999). https://doi.org/10.1023/A:1006115732620
Issue Date:
DOI: https://doi.org/10.1023/A:1006115732620