Abstract
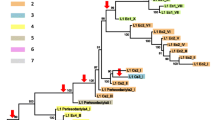
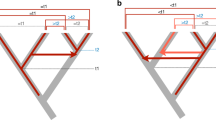
Since their discovery in family Bovidae (bovids), Bov-B LINEs, believed to be order-specific SINEs, have been found in all ruminants and recently also in Viperidae snakes. The distribution and the evolutionary relationships of Bov-B LINEs provide an indication of their origin and evolutionary dynamics in different species. The evolutionary origin of Bov-B LINE elements has been shown unequivocally to be in Squamata (squamates). The horizontal transfer of Bov-B LINE elements in vertebrates has been confirmed by their discontinuous phylogenetic distribution in Squamata (Serpentes and two lizard infra-orders) as well as in Ruminantia, by the high level of nucleotide identity, and by their phylogenetic relationships. The direction of horizontal transfer from Squamata to the ancestor of Ruminantia is evident from the genetic distances and discontinuous phylogenetic distribution of Bov-B LINE elements. The ancestral snake lineage (Boidae) has been recognized as a possible donor of Bov-B LINE elements to Ruminantia. The timing of horizontal transfer has been estimated from the distribution of Bov-B LINE elements in Ruminantia and the fossil data of Ruminantia to be 40–50mya. The phylogenetic relationships of Bov-B LINE elements from the various Squamata species agrees with that of the species phylogeny, suggesting that Bov-B LINE elements have been stably maintained by vertical transmission since the origin of Squamata in the Mesozoic era.
Similar content being viewed by others
References
Burke, W.D., H.S. Malik, W.C. Lathe III & T.H. Eickbush, 1998. Are retrotransposons long-term hitchhikers. Nature 392: 141–142.
Buntjer, J.B., J.A. Hoff & J.A. Lenstra, 1997. Artiodactyl interspersed DNA repeats in cetacean genomes. J. Mol. Evol. 45: 66–69.
Capy, P., D. Anxolabehere & T. Langin, 1994. The strange phylogenies of transposable elements: are horizontal transfers the only explanation? Trends Genet. 10: 7–12.
Clark, J.B. & M.G. Kidwell, 1997. A phylogenetic perspective on P transposable element evolution in Drosophila. Proc. Natl. Acad. Sci. USA 94: 11428–11433.
Deininger, P.L., M.A. Batzer, C.A. Hutchinson III & M.H. Edgell, 1992. Master genes in mammalian repetitive DNA amplification. Trends Genet. 8: 307–311.
Duncan, C.H., 1987. Novel Alu-type repeats in artiodactyls. Nucleic Acids Res. 15: 1340.
Eickbush, D.G. & T.H. Eickbush, 1995. Vertical transmission of the retrotransposable elements R1 and R2 during the evolution of the Drosophila melanogaster. Genetics 139: 671–684.
Felsenstein, J., 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17: 368–376.
Fitch, W.M., 1971. Toward defining the course of evolution: minimum change for a specific tree topology. Syst. Zool. 20: 406–416.
Gueiros-Filho, F.J. & S.M. Beverley, 1997. Trans-kingdom transposition of the Drosophila element mariner within the protozoan Leishmania. Science 276: 1716–1719.
Hamada, M., Y. Kido, M. Himberg, J.D. Reist, C. Ying et al., 1997. A newly isolated family of short interspersed repetitive elements (SINEs) in coregonid fishes (whitefish) with sequences that are almost identical to those of the Sma I family of repeats: possible evidence for the horizontal transfer of SINEs. Genetics 146: 355–367.
Houck, M.A., J.B. Clark, K.R. Peterson & M.G. Kidwell, 1991. Possible horizontal transfer of Drosophila genes by the mite Proctolaelaps regalis. Science 253: 1125–1129.
Hutchison III, C.A., S.C. Hardies, D.D. Loeb, W.R. Shehee & M.H. Edgell, 1989. LINEs and related retroposons, pp. 593–617 in Mobile DNA, edited by D.E. Berg, and M.M. Howe. American Society of Microbiology, Washington DC.
Jobse, C., J.B. Buntjer, N. Haagsma, H.J. Breukelman, J.J. Beintema et al., 1995. Evolution and recombination of bovine DNA repeats. J. Mol. Evol. 41: 277–283.
John, T.R., L.A. Smith & I.I. Kaiser, 1994. Genomic sequences encoding the acidic and basic subunits of Mojave toxin: unusually high sequence identity of non-coding regions. Gene 139: 229–234.
Kajikawa, M., K. Ohshima & N. Okada, 1997. Determination of the entire sequence of turtle CR1: the first open reading frame of the turtle CR1 element encodes a protein with a novel zinc finger motif. Mol. Biol. Evol. 14: 1206–1217.
Kidwell, M.G., 1993. Lateral transfer in natural populations of eukaryotes. Ann. Rev. Genet. 27: 235–256.
Kordiš D. & F. Gubenšek, 1995. Horizontal SINE transfer between vertebrate classes. Nat. Genet. 10: 131–132.
Kordiš D. & F. Gubenšek, 1996. Ammodytoxin C gene helps to elucidate the irregular structure of Crotalinae group II phospholipase PLA2 genes. Eur. J. Biochem. 240: 83–89.
Kordiš D. & F. Gubenšek, 1997. Bov-B long interspersed repeated DNA (LINE) sequences are present in Vipera ammodytes phospholipase A2 genes and in genomes of Viperidae snakes. Eur. J. Biochem. 246: 772–779.
Kordiš D. & F. Gubenšek, 1998. Unusual horizontal transfer of a long interspersed nuclear element between distant vertebrate classes. Proc. Natl. Acad. Sci. USA 95: 10704–10709.
Lenstra, J.A., J.A.F. van Boxtel, K.A. Zwaagstra & M. Schwerin, 1993. Short interspersed nuclear element (SINE) sequences of the Bovidae. Animal Genet. 24: 33–39.
Li, W.H., T. Gojobori & M. Nei, 1981. Pseudogenes as a paradigm of neutral evolution. Nature 292: 237–239.
Majewska, K., J. Szemraj, G. Plucienniczak, J. Jaworski & A. Plucienniczak, 1988. A new family of dispersed, highly repetitive sequences in bovine genome. Biochim. Biophys. Acta 949: 119–124.
Malik, H.S. & T.H. Eickbush, 1998. The RTE class of non-LTR retrotransposons is widely distributed in animals and is the origin of many SINEs. Mol. Biol. Evol. 15: 1123–1134.
Malik, H.S., W.D. Burke & T.H. Eickbush, 1999. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 16: 793–805.
Modi, W.S., D.S. Gallagher & J.E. Womack, 1996. Evolutionary histories of highly repeated DNA families among the Artiodactyla (Mammalia). J. Mol. Evol. 42: 337–349.
Nakashima, K., I. Nobuhisa, M. Deshimaru, M. Nakai, T. Ogawa et al., 1995. Accelerated evolution in the protein-coding regions is universal in crotalinae snake venom gland phospholipase A2 isozyme genes. Proc. Natl. Acad. Sci. USA 92: 5605–5609.
Nobukuni, T., M. Kobayashi, A. Omori, S. Ichinose, T. Iwanaga et al., 1997. An Alu-linked repetitive sequence corresponding to 280 amino acids is expressed in a novel bovine protein, but not in its human homologue. J. Biol. Chem. 272: 2801–2807.
Okada, N. & M. Hamada, 1997. The 3′ ends of of tRNA-derived SINEs originated from the 3′ end of LINEs: a new example from the bovine genome. J. Mol. Evol. 44(Suppl.): 52–56.
Okada, N., M. Hamada, I. Ogiwara & K. Ohshima, 1997. SINEs and LINEs share common 3′ sequences: a review. Gene 205: 229–243.
Pascale, E., C. Liu, E. Valle, K. Usdin & V. Furano, 1993. The evolution of long interspersed repeated DNA (L1, LINE 1) as revealed by the analysis of an ancient rodent L1 DNA family. J. Mol. Evol. 36: 9–20.
Robertson, H.M., 1993. The mariner transposable element is widespread in insects. Nature 362: 241–245.
Saitou, N. & M. Nei, 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425.
Shimamura, M., H. Abe, M. Nikaido, K. Ohshima & N. Okada, 1999. Genealogy of families of SINEs in cetaceans and artiodactyls: the presence of a huge superfamily of tRNA (Glu)-derived families of SINEs. Mol. Biol. Evol. 16: 1046–1060.
Smit, A.F.A., G. Toth, A.D. Riggs & J. Jurka, 1995. Ancestral mammalian-wide subfamilies of LINE-1 repetitive sequences. J. Mol. Biol. 246: 401–417.
Smit A.F.A., 1996. The origin of interspersed repeats in the human genome. Curr. Opin. Genet. Dev. 6: 743–748.
Szemraj, J., G. Plucienniczak, J. Jaworski & A. Plucienniczak, 1995. Bovine Alu-like sequences mediate transposition of a new site-specific retroelement. Gene 152: 261–264.
van De Peer, Y. & R. de Wachter, 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10: 569–570.
Zug, G.R., 1993. Herpetology: An Introductory Biology of Amphibians and Reptiles. Academic Press, San Diego.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Kordišs, D., Gubenšsek, F. Horizontal transfer of non-LTR retrotransposons in vertebrates. Genetica 107, 121–128 (1999). https://doi.org/10.1023/A:1004082906518
Issue Date:
DOI: https://doi.org/10.1023/A:1004082906518