Abstract
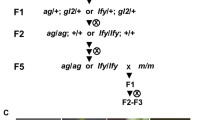
The stochastic variability of expression that is a characteristic of eukaryotic nuclear transgenes is often attributed to epigenetic mechanisms that are triggered by repetitive transgene locus structures and influenced by chromosomal position effects. In order to address the contribution of chromosomal position effects in the context of a fully sequenced genome, a novel set of transgene loci was established in the compact genome of Arabidopsis thaliana. Transgenes expressing GFP-tagged or GUS-tagged fusion proteins of Arabidopsis COP1 collectively displayed three types of gene silencing, which are distinguished by their developmental timing, gene dosage dependence, (post)transcriptional control, and extent of endogene co-suppression. Subsequently, the heritability of epistatic interactions between allelic and non-allelic transgene loci was investigated in light of both intrinsic transgene features, in particular T-DNA copy number per locus, and chromosomal insertion sites. The notion that chromosomal flanking sequences underlie the ability of transgenes to function as masters or targets of epigenetically heritable trans-silencing interactions was generally not favored by our data. Moreover, among single T-DNA loci at different chromosomal locations the great majority showed homozygosity-dependent posttranscriptional silencing. However, spontaneous silencing (in cis) may be promoted by a pericentromeric location. Instead, intrinsic transgene features correlated with all major aspects of silencing behavior tested.
Similar content being viewed by others
References
Al-Kaff, N.S., Covey, S.N., Kreike, M,M., Page, A.M., Pinder, R. and Dale, P.J. 1998. Transcriptional and posttranscriptional plant gene silencing in response to a pathogen. Science 279: 2113–2115.
Beclin, C., Boutet, S., Waterhouse, P. and Vaucheret, H. 2002. A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol. 12: 684–688.
Bender, J. 2001. A vicious cycle: RNA silencing and DNA methylation in plants. Cell 106: 129–132.
Birchler, J.A., Bhadra, M.P. and Bhadra, U. 2000. Making noise about silence: repression of repeated genes in animals. Curr. Opin. Genet. Dev. 10: 211–216.
Chandler, V.L., Eggleston, W.B. and Dorweiler, J.E. 2000. Paramutation in maize. Plant Mol. Biol. 43: 121–145.
Cogoni, C. 2001. Homology-dependent gene silencing mechanisms in fungi. Annu. Rev. Microbiol. 55: 381–406.
Day, C.D., Lee, E., Kobayashi, J., Holappa, L.D., Albert, H. and Ow, D.W. 2000. Transgene integration into the same chromosome location can produce alleles that express at a predictable level, or alleles that are differentially silenced. Genes Dev. 14: 2869–2880.
De Carvalho, F., Gheysem, G., Kushnir, S., Van Montagu, M., Inzé, D., and Castresana, C. 1992. Suppression of β-1,3-glucanase transgene expression in homozygous plants. EMBO J. 11: 2595–2602.
Dehio, C. and Schell, J. 1994. Identification of plant genetic loci involved in a posttranscriptional mechanism for meiotically reversible transgene silencing. Proc. Natl. Acad. Sci. USA 91: 5538–5542.
Dieguez, M.J., Vaucheret, H., Paszkowski, J. and Mittelsten Scheid, O. 1998. Cytosine methylation at CG and CNG sites is not a prerequisite for the initiation of transcriptional gene silencing in plants, but it is required for its maintenance. Mol. Gen. Genet. 259: 207–215.
Elmayan, T. and Vaucheret, H. 1996. Single copies of a 35S-driven transgene can undergo post-transcriptional silencing at each generation or can be transcriptionally inactivated in trans by a 35S silencer. Plant J. 9: 787–797.
Elmayan, T., Balzergue, S., Beon, F., Bourdon, V., Daubremet, J., Guenet, Y., Mourrain, P., Palauqui, J.C., Vernhettes, S., Vialle, T., Wostrikoff, K. and Vaucheret, H. 1998. Arabidopsis mutants impaired in cosuppression. Plant Cell 10: 1747–1758.
Fagard, M. and Vaucheret, H. 2000. (Trans)gene silencing in plants: how many mechanisms? Annu. Rev. Plant Physiol. Plant Mol. Biol. 51: 167–194.
Hamilton, A.J. and Baulcombe, D.C. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952.
Henikoff, S. 1998. Conspiracy of silencing among repeated transgenes. BioEssays 20: 532–535.
Hobbs, S.L., Warkentin, T.D. and DeLong, C.M. 1993. Transgene copy number can be positively or negatively associated with transgene expression. Plant Mol. Biol. 21: 17–26.
Hollick, J.B., Dorweiler, J.E. and Chandler, V.L. 1997. Paramutation and related allelic interactions. Trends Genet. 13: 302–308.
Iglesias, V., Moscone, E., Papp, I., Neuhuber, F., Michalowski, S., Phelan, T., Spiker, S., Matzke, M. and Matzke, A. 1997. Molecular and cytogenetic analysis of stably and unstably expressed transgene loci in tobacco. Plant Cell 9: 1251–1264.
Jakowitsch, J., Papp, I., Moscone, E.A., van der Winden, J., Matzke, M. and Matzke, A.J. 1999. Molecular and cytogenetic characterization of a transgene locus that induces silencing and methylation of homologous promoters in trans. Plant J. 17: 131–140.
Jeddeloh, J.A., Stokes, T.L. and Richards, E.J. 1999. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nature Genet. 22: 94–97.
Jones, L., Ratcliff, F. and Baulcombe, D.C. 2001. RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr. Biol. 11: 747–757.
Jorgensen, R.A., Cluster, P.D., English, J., Que, Q. and Napoli, C.A. 1996. Chalcone synthase cosuppression phenotypes in petunia flowers: comparison of sense vs. antisense constructs and singlecopy vs. complex T-DNA sequences. Plant Mol. Biol. 31: 957–973.
Jorgensen, R.A., Que, Q. and Stam, M. 1999. Do unintended antisense transcripts contribute to sense cosuppression in plants? Trends Genet. 15: 11–12.
Kim, T.H., Kim, B.H. and von Arnim, A.G. 2002. Repressors of photomorphogenesis. Int. Rev. Cytol. 220: 185–223.
Kooter, J.M., Matzke, M.A. and Meyer, P. 1999. Listening to the silent genes: transgene silencing, gene regulation and pathogen control. Trends Plant Sci. 4: 340–347.
Koprek, T., Rangel, S., McElroy, D., Louwerse, J.D., Williams-Carrier, R.E. and Lemaux, P.G. 2001. Transposon-mediated single-copy gene delivery leads to increased transgene expression stability in barley. Plant Physiol. 125: 1354–1362.
Lindroth, A.M., Cao, X., Jackson, J.P., Zilberman, D., McCallum, C.M., Henikoff, S. and Jacobsen, S.E. 2001. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292: 2077–2080.
Liu, Y.G., Mitsukawa, N., Oosumi, T. and Whittier, R.F. 1995. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8: 457–463.
Luff, B., Pawlowski, L. and Bender, J. 1999. An inverted repeat triggers cytosine methylation of identical sequences in Arabidopsis. Mol. Cell 3: 505–511.
Mallory, A.C., Ely, L., Smith, T.H., Marathe, R., Anandalakshmi, R., Fagard, M., Vaucheret, H., Pruss, G., Bowman, L. and Vance, V.B. 2001. HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell 13: 571–583.
Martienssen, R. 1996. Epigenetic phenomena: paramutation and gene silencing in plants. Curr. Biol. 6: 810–813.
Martienssen, R. 1998. Chromosomal imprinting in plants. Curr. Opin. Genet. Dev. 8: 240–244.
Martinez-Perez, E., Shaw, P.J. and Moore, G. 2000. Polyploidy induces centromere association. J. Cell Biol. 148: 233–238.
Matzke, A.J. and Matzke, M.A. 1998. Position effects and epigenetic silencing of plant transgenes. Curr. Opin. Plant Biol. 1: 142–148.
McNellis, T., von Arnim, A.G. and Deng, X.W. 1994. Overexpression of Arabidopsis COP1 results in partial suppression of light-mediated development: evidence for a light-inactivable repressor of photomorphogenesis. Plant Cell 6: 1391–1400.
Meins, F. Jr. 2000. RNA degradation and models for posttranscriptional gene-silencing. Plant Mol. Biol. 43: 261–273.
Mette, M.F., van der Winden, J., Matzke, M.A. and Matzke, A.J. 1999. Production of aberrant promoter transcripts contributes to methylation and silencing of unlinked homologous promoters in trans. EMBO J. 18: 241–248.
Mette, M.F., Aufsatz, W., van der Winden, J., Matzke, M.A. and Matzke, A.J. 2000. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 19: 5194–5201.
Meza, T.J., Stangeland, B., Mercy, I.S., Skarn, M., Nymoen, D.A., Berg, A., Butenko, M.A., Hakelien, A.M., Haslekas, C., Meza-Zepeda, L.A. and Aalen, R.B. 2002. Analyses of single-copy Arabidopsis T-DNA-transformed lines show that the presence of vector backbone sequences, short inverted repeats and DNA methylation is not sufficient or necessary for the induction of transgene silencing. Nucl. Acids Res. 30: 4556–4566.
Meyer, P., Heidmann, I. and Niedenhof, I. 1993. Differences in DNA-methylation are associated with a paramutation phenomenon in transgenic petunia. Plant J. 4: 89–100.
Miséra, S., Müller, A.J., Weiland-Heidecker, U. and Jürgens, G. 1994. The FUSCA genes of Arabidopsis: negative regulators of light responses. Mol. Gen. Genet. 244: 242–252.
Mitsuhara, I., Shirasawa-Seo, N., Iwai, T., Nakamura, S., Honkura, R. and Ohashi, Y. 2002. Release from post-transcriptional gene silencing by cell proliferation in transgenic tobacco plants: possible mechanism for noninheritance of the silencing. Genetics 160: 343–352.
Mittelsten Scheid, O., Jakovleva, L., Afsar, K., Maluszynska, J. and Paszkowski, J. 1996. A change of ploidy can modify epigenetic silencing. Proc. Natl. Acad. Sci. USA 93: 7114–7119.
Mlynarova, L., Loonen, A., Mietkiewska, E., Jansen, R.C. and Nap, J.P. 2002. Assembly of two transgenes in an artificial chromatin domain gives highly coordinated expression in tobacco. Genetics 160: 727–740.
Panavas, T., Weir, J. and Walker, E.L. 1998. The structure and paramutagenicity of the R-marbled haplotype of Zea mays. Genetics 153: 979–991.
Park, Y.D., Papp, I., Moscone, E.A., Iglesias, V.A., Vaucheret, H., Matzke, A.J. and Matzke, M.A. 1996. Gene silencing mediated by promoter homology occurs at the level of transcription and results in meiotically heritable alterations in methylation and gene activity. Plant J. 9: 183–194.
Pikaard, C.S. 1999. Nucleolar dominance and silencing of transcription. Trends Plant Sci. 4: 478–483.
Pröls, F. and Meyer, P. 1992. The methylation patterns of chromosomal integration regions influence gene activity of transferred DNA in Petunia hybrida. Plant J. 2: 465–475.
Que, Q. and Jorgensen, R.A. 1998. Homology-based control of gene expression patterns in transgenic petunia flowers. Dev. Genet. 22: 100–109.
Qin, H. and von Arnim, A.G. 2002. Epigenetic history of an Arabidopsis trans-silencer locus and a test for relay of trans-silencing activity. BMC Plant Biol. 2: 11.
Restrepo, M.A., Freed, D.D. and Carrington, J.C. 1990. Nuclear transport of plant potyviral proteins. Plant Cell 2: 987–998.
Sharp, P.A. 2001. RNA interference-2001. Genes Dev. 15: 485–490.
Sidorenko, L.V. and Peterson, T. 2001. Transgene-induced silencing identifies sequences involved in the establishment of paramutation of the maize p1 gene. Plant Cell 13: 319–335.
Sijen, T., Vijn, I., Rebocho, A., van Blokland, R., Roelofs, D., Mol, J.N. and Kooter, J.M. 2001. Transcriptional and posttranscriptional gene silencing are mechanistically related. Curr. Biol. 11: 436–440.
Smith, N.A., Singh, S.P., Wang, M.B., Stoutjesdijk, P.A., Green, A.G. and Waterhouse, P.M. 2000. Total silencing by intronspliced hairpin RNAs. Nature 407: 319–320.
Stacey, M.G., Hicks, S.N. and von Arnim, A.G. 1999. Discrete domains mediate the light-responsive nuclear and cytoplasmic localization of Arabidopsis COP1. Plant Cell 11: 349–364.
Stacey, M.G., Kopp, O.R., Kim, T.-H. and von Arnim, A.G. 2000. Modular domain structure of Arabidopsis COP1: reconstitution of activity by fragment complementation and mutational analysis of a nuclear localization signal in planta. Plant Physiology 124: 979–990 (2000).
Stam, M., Belele, C., Dorweiler, D.E. and Chandler, V.L. 2002. Differential chromatin structure within a tandem array 100kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 16: 1906–1918.
Stam, M., Viterbo, A., Mol, J.N. and Kooter, J.M. 1998. Positiondependent methylation and transcriptional silencing of transgenes in inverted T-DNA repeats: implications for posttranscriptional silencing of homologous host genes in plants. Mol. Cell. Biol. 18: 6165–6177.
ten Lohuis, M., Müller, A., Heidmann, I., Niedenhof, I. and Meyer, P. 1995. A repetitive DNA fragment carrying a hot spot for de novo DNA methylation enhances expression variegation in tobacco and petunia. Plant J. 8: 919–932.
Vaucheret, H., Elmayan, T., Thierry, D., van der Geest, A., Hall, T., Conner, A.J., Mlynarova, L. and Nap, J.P. 1998. Flank matrix attachment regions (MARs) from chicken, bean, yeast or tobacco do not prevent homology-dependent trans-silencing in transgenic tobacco plants. Mol. Gen. Genet. 259: 388–392.
Vaucheret, H., Nussaume, L., Palauqui, J.C., Quilléré, I. and Elmayan, T. 1997. A transcriptionally active state is required for post-transcriptional silencing (cosuppression) of nitrate reductase host genes and transgenes. Plant Cell 9: 1495–1504.
von Arnim, A.G. and Deng, X.W. 1994. Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cellspecific regulation of its nucleocytoplasmic partitioning. Cell 79: 1035–1045.
von Arnim, A.G., Deng, X.W. and Stacey, M.G. 1998. Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221: 35–43.
Walker, E.L. 1998. Paramutation of the r1 locus of maize is associated with increased cytosine methylation. Genetics 148: 1973–1981.
Walker, E.L. and Panavas, T. 2001. Structural features and methylation patterns associated with paramutation at the r1 locus of Zea mays. Genetics 159: 1201–1215.
Wang, M.B. and Waterhouse, P.M. 2000. High-efficiency silencing of a beta-glucuronidase gene in rice is correlated with repetitive transgene structure but is independent of DNAmethylation. Plant Mol. Biol. 43: 67–82.
Wassenegger, M. 2000. RNA-directed DNA methylation. PlantMol. Biol. 43: 203–220.
Waterhouse, P.M., Wang, M.B. and Finnegan, E.J. 2001. Role of short RNAs in gene silencing. Trends Plant Sci. 6: 297–301.
Ye, F. and Signer, E.R. 1996. RIGS (repeat-induced gene silencing) in Arabidopsis is transcriptional and alters chromatin configuration. Proc. Natl. Acad. Sci. USA 93: 10881–10886.
Zuker, M., Mathews, D.H. and Turner, D.H. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In: J. Barciszewski and B.F.C. Clark (Eds.) RNA Biochemistry and Biotechnology, NATO ASI Series, Kluwer Academic Publishers, pp. 11–43.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Qin, H., Dong, Y. & von Arnim, A.G. Epigenetic interactions between Arabidopsis transgenes: characterization in light of transgene integration sites. Plant Mol Biol 52, 217–231 (2003). https://doi.org/10.1023/A:1023941123149
Issue Date:
DOI: https://doi.org/10.1023/A:1023941123149