Abstract
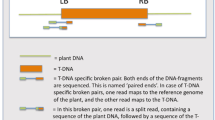
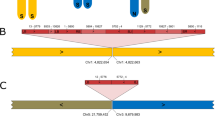
T-DNA flanking sequences were isolated from 112 Arabidopsis thaliana single-copy T-DNA lines and sequence mapped to the chromosomes. Even though two T-DNA insertions mapped to a heterochromatic domain located in the pericentromeric region of chromosome I, expression of reporter genes was detected in these transgenic lines. T-DNA insertion did not seem to be biased toward any of Arabidopsis' five chromosomes. The observed distribution of T-DNA copies in intergenic sequence versus gene sequence (i.e. 5′-upstream regions, coding sequences and 3′-downstream regions) appeared randomly. An evaluation of T-DNA insertion frequencies within gene sequence revealed that integration into 5′-upstream regions occurred more frequently than expected, whereas insertions in coding sequences (exons and introns) were found less frequently than expected based on random distribution predictions. In the majority of cases, single-copy T-DNA insertions were associated with small or large rearrangements such as deletions and/or duplications of target site sequences, deletions and/or duplications of T-DNA sequences, and gross chromosomal rearrangements such as translocations. The accuracy of integration was similarly high for both left- and right-border sequences. These results may be called upon when making detailed molecular analyses of transgenic plants or T-DNA induced mutants.
Similar content being viewed by others
References
Altschul, S.F., Gish, W., Miller, W., Myers, E.W. and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410.
Arabidopsis Genome Initiative. 2000. Sequence and analysis of the flowering plant Arabidopsis thaliana. Nature 408: 796–815.
Azpiroz-Leehan, R. and Feldmann K.A. 1997. T-DNA insertion mutagenesis in Arabidopsis: going back and forth. Trends Genet. 13: 152–156.
Bechtold, N., Ellis, J. and Pelletier, G. 1993. In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad. Sci. Paris 316: 1194–1199.
Bundock, P., van Attikum, H., den Dulk-Ras, A. and Hooykaas, P.J. 2002. Insertional mutagenesis in yeasts using T-DNA from Agrobacterium tumefaciens. Yeast 19: 529–536.
Castle, L.A., Errampalli, D., Atherton, T.L., Franzmann, L.H., Yoon, E.S., Meinke, D.W. 1993. Genetic and molecular characterization of embryonic mutants identified following seed transformation in Arabidopsis. Mol. Gen. Genet. 241: 504–514.
Clough, S.J. and Bent, A.F. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743.
Cold Spring Harbor Laboratory, Washington University Genome Sequencing Center, and PE Biosystems Arabidopsis Sequencing Consortium. 2000. The complete sequence of a heterochromatic island from a higher eukaryote. Cell 100: 377–386.
Dellaporta, S.L., Wood, J. and Hicks, J.B. 1983. A plant DNA minipreparation: version II. Plant Mol. Biol. Rep. 1: 19–21.
De Neve, M., De Buck, S., Jacobs, A., Van Montagu, M. and Depicker, A. 1997. T-DNA integration patterns in cotransformed plant cells suggest that T-DNA repeats originate from co-integration of separate T-DNAs. Plant J. 11: 15–29.
Dujon, B. 1996. The yeast genome project: what did we learn? Trends Genet. 12: 263–270.
Gheysen, G., Van Montagu, M. and Zambyski, P. 1987. Integration of Agrobacterium tumefaciens transfer DNA (T-DNA) involves rearrangements of target plant DNA sequences. Proc. Natl. Acad. Sci. USA 84: 6169–6173.
Gheysen, G., Villarroel, R. and Van Montagu, M. 1991. Illegitimate recombination in plants: a model for T-DNA integration. Genes Dev. 5: 287–297.
Haupt, W., Fischer, T.C., Winderl, S., Fransz, P. and Torres-Ruiz, R.A. 2001. The centromere 1 (CEN1) region of Arabidopsis thaliana: architecture and functional impact of chromatin. Plant J. 27: 285–296.
Herman, L., Jacobs, A., Van Montagu, M. and Depicker, A. 1990. Plant chromosome/marker gene fusion assay for study of normal and truncated T-DNA integration events. Mol. Gen. Genet. 224: 248–256.
Höfgen, R. and Willmitzer, L. 1988. Storage of competent cells for Agrobacterium transformation. Nucl. Acids Res. 16: 9877.
Iglesias, V.A., Moscone, E.A., Papp, I., Neuhuber, F., Michalowski, S., Phelan, T., Spiker, S., Matzke, M. and Matzke, A.J. 1997. Molecular and cytogenetic analyses of stably and unstably expressed transgene loci in tobacco. Plant Cell9: 1251–1264.
Kertbundit, S., De Greve, H., Deboeck, F., Van Montagu, M. and Hernalsteens, J.P. 1991. In vivo random β-glucuronidase gene fusions in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 88:5212–5216.
Koncz, C., Martini, N., Mayerhofer, R., Koncz-Kalman, Z., Korber, H., Redei, G.P. and Schell, J. 1989. High-frequency T-DNAmediated gene tagging in plants. Proc. Natl. Acad. Sci. USA 86: 8467–8471.
Kononov, M.E., Bassuner, B. and Gelvin, S.B. 1997. Integration of T-DNA binary vector 'backbone' sequences into the tobacco genome: evidence for multiple complex patterns of integration. Plant J. 11: 945–957.
Laufs, P., Autran, D. and Traas, J., 1999. A chromosomal paracentric inversion associated with T-DNA integration in Arabidopsis. Plant J. 18: 131–139</del>.
Lessl, M. and Lanka, E. 1994. Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell 77: 321–324.
Martineau, B., Voelker, T.A. and Sanders, R.A. 1994. On defining T-DNA. Plant Cell 6: 1032–1033.
Mathur, J., Szabados, L., Schaefer, S., Grunenberg, B., Lossow, A., Jonas-Straube, E., Schell, J., Koncz, C. and Koncz-Kalman, Z. 1998. Gene identification with sequenced T-DNA tags generated by transformation of Arabidopsis cell suspension. Plant J. 13: 707–716.
Mayerhofer, R., Koncz-Kalman, Z., Nawrath, C., Bakkeren, G., Crameri, A., Angelis, K., Redei, G.P., Schell, J., Hohn, B. and Koncz, C. 1991. T-DNA integration: a mode of illegitimate recombination in plants. EMBO J. 10: 697–704.
McKinney, E.C., Ali, N., Traut, A., Feldmann, K.A., Belostotsky, D.A., McDowell, J.M., Meagher, R.B. 1995. Sequence-based identification of T-DNA insertion mutations in Arabidopsis: actin mutants act2–1 and act4–1. Plant J. 8: 613–622.
Meissner, R.C., Jin, H., Cominelli, E., Denekamp, M., Fuertes, A., Greco, R., Kranz, H.D., Penfield, S., Petroni, K., Urzainqui, A., Martin, C., Paz-Ares, J., Smeekens, S., Tonelli, C., Weisshaar, B., Baumann, E., Klimyuk, V., Marillonnet, S., Patel, K., Speulman, E., Tissier, A.F., Bouchez, D., Jones, J.D., Pereira, A., Wisman, E. and Bevan, M. 1999. Function search in a large transcription factor gene family in Arabidopsis: assessing the potential of reverse genetics to identify insertional mutations in R2R3 MYB genes. Plant Cell 11: 1827–1840.
Nacry, P., Camilleri, C., Courtial, B., Caboche, M. and Bouchez, D. 1998. Major chromosomal rearrangements induced by T-DNA transformation in Arabidopsis. Genetics 149: 641–650.
Neuhuber, F., Park, Y.D., Matzke, A.J. and Matzke, M.A. 1994. Susceptibility of transgene loci to homology-dependent gene silencing. Mol. Gen. Genet. 244: 230–241.
Ochman, H., Gerber, A.S. and Hartl, D.L. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120: 621–623.
Ohba, T., Yoshioka, Y., Machida, C. and Machida, Y. 1995. DNA rearrangement associated with the integration of T-DNA in tobacco: an example for multiple duplications of DNA around the integration target. Plant J. 7: 157–164.
Parinov, S., Sevugan, M., De, Y., Yang, W.C., Kumaran, M. and Sundaresan, V. 1999. Analysis of flanking sequences from dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell 11: 2263–2270.
Raina, S, Mahalingam, R., Chen, F. and Fedoroff, N. 2002. A collection of sequenced and mapped Ds transposon insertion sites in Arabidopsis thaliana. Plant Mol. Biol. 50: 93–110.
Rossi, L., Hohn, B. and Tinland, B. 1996. Integration of complete transferred DNA units is dependent on the activity of virulence E2 protein of Agrobacterium tumefaciens. Proc Natl. Acad. Sci. USA 9: 126–130.
Sambrook, J., Fritsch, E.F. and Maniatis, T. 1989. Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, NY.
Sussman, M.R., Amasino, R.M., Young, J.C., Krysan, P.J. and Austin-Phillips, S. 2000. The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol. 124: 1465–1467.
Szabados, L., Kovacs, I., Oberschall, A., Abraham, E., Kerekes, I., Zsigmond, L., Nagy, R., Alvarado, M., Krasovskaja, I., Gal, M., Berente, A., Redei, G.P., Haim, A.B. and Koncz, C. 2002. Distribution of 1000 sequenced T-DNA tags in the Arabidopsis genome. Plant J. 32: 233–242.
Tatusova, T.A. and Madden, T.L. 1999. BLAST 2 sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174: 247–250.
Tax, F.E. and Vernon, D.M. 2001. T-DNA-associated duplication/ translocations in Arabidopsis. Implications for mutant analysis and functional genomics. Plant Physiol. 126: 1527–1538.
Thomas, C.M., Jones, D.A., English, J.J., Carroll, B.J., Bennetzen, J.L., Harrison, K., Burbidge, A., Bishop, G.J. and Jones, J.D. 1994. Analysis of the chromosomal distribution of transposoncarrying T-DNAs in tomato using the inverse polymerase chain reaction. Mol. Gen. Genet. 242: 573–585.
Tinland, B., Schoumacher, F., Gloeckler, V., Bravo-Angel, A.M. and Hohn, B. 1995. The Agrobacterium tumefaciens virulence D2 protein is responsible for precise integration of T-DNA into the plant genome. EMBO J. 14: 3585–3595.
Tinland, B. 1996. The integration of T-DNA into plant genomes. Trends Plant Sci. 1: 178–184.
Tissier, A.F., Marillonnet, S., Klimyuk, V., Patel, K., Torres, M.A., Murphy, G. and Jones, J.D. 1999. Multiple independent defective suppressor-mutator transposon insertions in Arabidopsis: a tool for functional genomics. Plant Cell 11: 1841–1852.
Valvekens, D., Van Montagu, M. and Van Lijsebettens, M. 1988. Agrobacterium tumefaciens-mediated transformation of Arabidopsis root explants using kanamycin selection. Proc. Natl. Acad. Sci. USA 85: 5536–5540.
van der Graaff, E., den Dulk-Ras, A. and Hooykaas, P.J. 1996. Deviating T-DNA transfer from Agrobacterium tumefaciens to plants. Plant Mol. Biol. 31: 677–681.
Wakimoto, B.T. 1998. Beyond the nucleosome: epigenetic aspects of position-effect variegation in Drosophila. Cell 93: 321–324.
Wenck, A., Czakó, M., Kanevski, I., Márton, L. 1997. Frequent collinear long transfer of DNA inclusive of the whole binary vector during Agrobacterium-mediated transformation. Plant Mol. Biol. 34: 913–922.
Zupan, J. and Zambryski, P. 1997. The Agrobacterium DNA transfer complex. Crit. Rev. Plant Sci. 16: 279–295.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Forsbach, A., Schubert, D., Lechtenberg, B. et al. A comprehensive characterization of single-copy T-DNA insertions in the Arabidopsis thaliana genome. Plant Mol Biol 52, 161–176 (2003). https://doi.org/10.1023/A:1023929630687
Issue Date:
DOI: https://doi.org/10.1023/A:1023929630687