Abstract
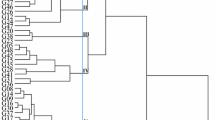
Twenty-two accessions belonging to seven species of Sesbania Scop. (S. emerus, S. exasperata, S. grandiflora, S. rostrata, S. sesban, S. tetrapteraand S. virgata), were evaluated in order to characterize the isozyme electrophoretic patterns and to estimate their genetic variability and outcrossing rates. Eight isozyme systems were used: acid phosphatase (ACP), isocitrate dehydrogenase (IDH), phosphoglucomutase (PGM), malate dehydrogenase (MDH), phosphoglucoisomerase (PGI), glutamate oxaloacetate transaminase (GOT), peroxidase (PRX) and catalase (CAT), with two gel/electrode buffers and distilled water as the extraction buffer. Cotyledons were chosen for enzyme extraction. The accessions were compared considering the presence/absence of bands for each system. A dendrogram was obtained using the Jaccard similarity index and the UPGMA clustering method, classifying the accessions into eight groups, evidencing both inter- and intraspecific variability. Annual species were classified in one major group separate from the perennials (S. grandiflora, S. sesban, S. virgata), which were allocated into two other main groups. S. virgata accessions (subgenus Daubentonia) were clearly separated from the other species. Isozyme banding patterns for the accessions are presented for each system. A high incidence of intraspecific monomorphism was observed. Interpretation in terms of loci and alleles was made for S. sesbanand S. virgataaccessions, which presented polymorphism for the systems PGI, GOT (S. sesban) and MDH (S. virgata). The average expected heterozygosity estimates varied from 0.0 to 0.231 for S. sesban and 0.0 to 0.20 for S. virgata. A mean number of 2.0 alleles per locus was observed for both species. Population multilocus outcrossing rate (t m) of 0.62 was estimated for S. sesban, indicating a partial allogamy mode of reproduction. Individual families presented multilocus rates varying from 0.00 to 1.00, indicating both self-pollination and cross-pollination.
Similar content being viewed by others
References
Alfenas, A.C., I. Peters, W. Brune & G.C. Passador, 1991. Eletroforese de proteínas e isoenzimas de fungos e essências florestais, 2nd ed Universidade Federal de Viçosa, Viçosa.
Brewbaker, J.L., 1990. Breeding systems and genetic improvement of perennial sesbanias. In: Macklin, B. & D.O. Evans (eds.), Perennial Sesbania Species in Agroforestry Systems, pp. 39–44, Nitrogen Fixing Tree Association, Waimanalo.
Brewbaker, J.L., M.D. Upadhya, J. Makinen & T. MacDonald, 1968. Isoenzyme polymorphism in flowering plants: III. Gel electrophoretic methods and applications. Physiol. Plant 21: 930–940.
Brewer, G.J. & C.F. Sing, 1970. An Introduction to Isozyme Techniques. Academic Press, New York.
Carulli, J.P. & D.E. Fairbrothers, 1988. Allozyme variation in three eastern United States species of Aeschynomene (Fabaceae), including the rare A. virginica. Syst. Bot. 13: 559–566.
Chamberlain, J.R., C.E. Hughes & N.W. Galwey, 1996. Patterns of isozyme variation in the Leucaena shannonii alliance (Leguminosae:Mimosoideae). Silvae Genet. 45: 1–7.
Cruz, C.D & A.J. Regazzi, 1994. Modelos biométricos aplicados ao melhoramento genético. Universidade Federal de Viçosa, Viçosa.
Evans, D.O., 1988. Perennial species of Sesbania. Nitrogen Fixing Tree Res. Rep. 6: 73–75.
Evans, D.O., 1990. What is Sesbania? Botany, taxonomy, plant geography, and natural history of the perennial members of the genus. In: Macklin, B. & D.O. Evans (eds.), Perennial Sesbania Species in Agroforestry Systems, pp. 5–16, Nitrogen Fixing Tree Association, Waimanalo.
Gutteridge, R.C., 1994. The perennial Sesbania species. In: Gutteridge, R.C. & H.M. Shelton (eds.), Forage Tree Legumes in Tropical Agriculture, pp. 49–64, CAB International, Wallingford.
Haldane, J.B.S., 1954. An exact test for randomness of mating. J. Genet. 52: 631–635.
Hamrick, J.L., 1990. Isozyme and the analysis of genetic structure in plant populations. In: Soltis, D.E. & P.S. Soltis (eds.), Isozymes in Plant Biology, pp.87–105, Chapman and Hall, London.
Harris, S.A., C.E. Hughes, R.J. Abbott & R. Ingram, R., 1994. Genetic variation in Leucaena leucocephala (Lam.) de Wit. (Leguminosae:Mimosoideae). Silvae Genet. 43: 159–167.
Heering, J.H., 1994. The reproductive biology of three perennial Sesbania species (Leguminosae). Euphytica 74: 143–148.
Heering, J.H., J.D. Reed & J. Hanson, 1996. Differences in Sesbania sesban accessions in relation to their phenolic concentration and HPLC fingerprints. J. Sci. Fd Agric. 71: 92–98.
Jain, S.K. & R.S. Singh, 1979. Population biology of Avena. VII. Allozyme variation in relation to the genome analysis. Bot. Gaz. 140: 356–362.
Kumar, S., K. Tamura & M. Nei, 1993. MEGA — molecular evolutionary analysis. Institute of Molecular Evolutionary Genetics, Pennsylvania State University, PA.
Lovato, M.B. & P.S. Martins, 1997. Genetic variability in salt tolerance during germination of Stylosanthes humilis H.B.K. and association between salt tolerance and isozymes. Rev. Brasil. Genet. 20: 435–441.
Marcon, G., 1988. Estrutura genética de populações de Stylosanthes humilis H.B.K. (Leguminosae) de tres regiões ecogeográficas do Estado de Pernambuco. Ph.D. Thesis. Escola Superior de Agricultura ‘Luiz de Queiroz’, University of São Paulo, Piracicaba, Brasil.
Miller, M.P., 1997. Tools for population genetic analyses (TFPGA). Version 1.3. Northern Arizona University, Flagstaff, AZ.
Monteiro, R., 1984. Taxonomic studies on Brazilian legumes with forage potential: Sesbania, Lupinus. Ph.D. Thesis. University of St. Andrews, St. Andrews, UK.
Ndoye, I., K. Tomekpe, B. Dreyfus & Y.R. Dommergues, 1990. Sesbania and Rhizobium symbiosis: nodulation and nitrogen fixation. In: Macklin, B. & D.O. Evans (eds.), Perennial Sesbania Species in Agroforestry Systems, pp. 31–38, Nitrogen Fixing Tree Association, Waimanalo.
Nei, M., 1973. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 70: 3321–3323.
Nei, M., 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89: 583–590.
Penteado, M.I. de O., L.E. Sáenz de Miera & M. Pérez de la Veja, 1996. Genetic resources of Centrosema spp.: genetic changes associated to the handling of an active collection. Genet. Resour. Crop Evolution 43: 85–90.
Penteado, M.I. de O., P. Garcia & M. Pérez de la Veja, 1997. Isozyme markers and genetic variability in three species of Centrosema (Leguminosae). Rev. Brasil. Genet. 20: 443–452.
Plumb, V.E., C.D. Wood, C. Gay, J. Hanson & J.H. Heering, 1996. The use of HPLC-derived phenolic profiles as a means of classifying Sesbania accessions. J. Sci. Fd Agric. 70: 481–486.
Reis, M.S. dos, 1996. Distribuição e dinâmica da variabilidade genética em populações naturais de palmiteiro (Euterpe edulis Martius). Ph.D. Thesis. Escola Superior de Agricultura ‘Luiz de Queirozd’, University of São Paulo, Piracicaba, Brasil.
Ritland, K., 1996. Multilocus mating system program MLTR. Version 1.1. University of Toronto, Toronto.
Ritland, K. & S. K. Jain, 1981. A model for the estimation of outcrossing rate and gene frequencies using n independent loci. Heredity 47: 35–52.
Saraswati, R., T. Matoh, T. Sasai, P. Phupaibul, T.A. Lumpkin, M. Kobayashi & J. Sekiya, 1993. Identification of Sesbania species from electrophoretic patterns of seed proteins. Trop. Agric. 70: 282–285.
Scandalios, J.G., 1965. Genetic isozyme variations in Zea mays. Ph.D. Thesis. University of Hawaii, Honolulu.
Simpson, M.J.A. & L.A. Withers, 1986. Characterization of plant genetic resources using isozyme electrophoresis: a guide to the literature. International Board For Plant Genetic Resources, Rome.
Soltis, D.E. & P.E. Soltis, 1990. Isozymes in Plant Biology. Chapman and Hall, London.
Stat Soft Inc., 1996. Statistica forWindows. Release 5.1. (software).
Soltis, D.E., C.H. Haufler, D.C. Darrow & G.S. Gastony, 1983. Starch gel electrophoresis of ferns: a compilation of grinding buffers, gel and electrode buffers, and staining schedules. Am. Fern J. 73: 9–27.
Veasey, E.A., E.A. Schammass, R. Vencovsky, P.S. Martins & G. Bandel, 1999. Morphological and agronomical characterization and estimates of genetic parameters of Sesbania Scop. (Leguminosae) accessions. Genet. Mol. Biol. 22: 81–93.
Veasey, E.A., E.A. Schammass, R. Vencovsky, P.S. Martins & G. Bandel, 2001. Germplasm characterization of Sesbania accessions based on multivariate analyses. Genet. Resour. Crop Evolution 48: 79–90.
Wendel, J.F. & N.F. Weeden, 1990. Visualization and interpretation of plant isozymes. In: Soltis, D.E. & P.S. Soltis (eds.), Isozymes in Plant Biology, pp. 5–45, Chapman and Hall, London.
Wright, S., 1978. Evolution and the Genetics of Populations, Vol. 4. University of Chicago Press, Chicago.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Veasey, E.A., Vencovsky, R., Sodero Martins, P. et al. Germplasm characterization of Sesbania accessions based on isozyme analyses. Genetic Resources and Crop Evolution 49, 449–462 (2002). https://doi.org/10.1023/A:1020998913573
Issue Date:
DOI: https://doi.org/10.1023/A:1020998913573