Abstract
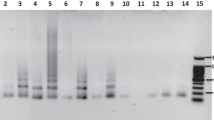
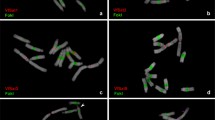
Telomeres, DNA–protein structures, are important elements of the eukaryotic chromosome. Telomeric regions of the majority of higher plants contain heptanucleotides TTTAGGG arranged into a tandem repeat. However, some taxa have no such repeats. These are some species of Liliaceae and Alliaceae. For example, terminal regions of chromosomes of bunching onion (Allium fistulosum) contain satellite DNA whose unit repeats are 380 bp in length, and the short arm of its chromosome 8 contains rDNA repeats. This study deals with the terminal heterochromatin and organization of the satellite repeat in A. fistulosum. Fluorescent in situ hybridization (FISH) was used to locate the satellite DNA on chromosomes and on extended DNA of A. fistulosum.Nonsatellite DNA was found in the structure of telomeric repeat. Polymerase chain reaction (PCR) and Southern hybridization were used for analysis of terminal heterochromatin. Various rearrangements were found in the satellite repeat. The roles of retrotransposons and microsatellites in the formation of terminal heterochromatin are discussed.
Similar content being viewed by others
REFERENCES
Zakian, V.A., Telomeres: Beginning to Understand the End, Science, 1995, vol. 270, pp. 1601-1607.
Heslop-Harrison, J.S., Leitch, A.R., and Schwarzacher, T., The Physical Organization of Interphase Nuclei, The Chromosome, Heslop-Harrison, J.S. and Flavell, R., Eds., Oxford: BIOS Scientific, 1993, pp. 221-232.
Fuchs, J., Brandes, A., and Schubert, I., Telomere Sequence Localization and Karyotype Evolution in Higher Plants, Plant Syst. Evol., 1995, vol. 196, pp. 227-241.
Richards, E.J. and Ausubel, F.M., Isolation of a Higher Eukaryotic Telomere from Arabidopsis thaliana, Cell (Cambridge, Mass.), 1988, vol. 53, pp. 127-136.
Pich, U., Fuchs, J., and Schubert, I., How Do Alliaceae Stabilize Their Chromosome Ends in the Absence of TTTAGGG Sequences?, Chromosome Res., 1996, vol. 4, pp. 207-213.
Irifune, K., Hirai, K., Zheng, J., et al., Nucleotide Sequence of a Highly Repeated DNA Sequence and Its Chromosomal Localization in Allium fistulosum, Theor. Appl. Genet., 1995, vol. 90, pp. 312-316.
Pich, U. and Schubert, I., Terminal Heterochromatin and Alternative Telomeric Sequences in Allium cepa, Chromosome Res., 1998, vol. 6, pp. 315-321.
Pich, U., Fritsch, R., and Shubert, I., Closely Related Allium Species (Alliaceae) Share a Very Similar Satellite Sequence, Plant Syst. Evol., 1996, vol. 202, pp. 255-264.
Biessmann, H. and Mason, J.M., Genetics and Molecular Biology of Telomeres, Adv. Genet., 1992, vol. 30, pp. 185-249.
Lopez, C.C., Nielsen, L., and Edstrom, J.-E., Terminal Long Tandem Repeats in Chromosomes from Chironomus pallidivittatus, Mol. Cell. Biol., 1996, vol. 16, no. 7, pp. 3285-3290.
Bernatzky, R. and Tanksley, S.D., Genetics of Actin-Related Sequence in Tomato, Theor. Appl. Genet., 1986, vol. 72, pp. 314-324.
Karlov, G.I., Khrustaleva, L.I., Lim, K.B., and Van Tuyl, J.M., Homoeologous Recombination in 2n Gametes Producing Interspecific Hybrids of Lilium (Liliaceae) Studied by Genomic In Situ Hybridization (GISH), Genome, 1999, vol. 42, pp. 681-686.
Ganal, M.W., Lapitan, N.L.V., and Tanksley, S.D., Macrostructure of the Tomato Telomeres, Plant Cell, 1992, vol. 3, pp. 87-94.
Wu, K.-S. and Tanksley, S.D., Genetic and Physical Mapping of Telomeres and Microsatellites of Rice, Plant Mol. Biol., 1993, vol. 22, pp. 861-872.
Wu, K.-S. and Tanksley, S.D., PFGE Analysis of the Rice Genome: Estimation of Fragment Sizes, Organization of Repetitive Sequences and Relationship between Genetic and Physical Distances, Plant Mol. Biol., 1993, vol. 23, pp. 243-254.
Roder, M.S., Lapitan, N.L.V., Sorrells, M.E., and Tanksley, S.D., Genetic and Physical Mapping of Barley Telomeres, Mol. Gen. Genet., 1993, vol. 238, pp. 294-303.
Verchinin, A.V., Schwazacher, T., and Heslop-Harrison, J.S., The Large-Scale Genomic Organization of Repetitive DNA Families at the Telomeres of Rye Chromosomes, Plant Cell, 1995, vol. 7, pp. 1823-1833.
Narayan, R.K.J., Constraints upon the Organization and Evolution of Chromosome in Allium, Theor. Appl. Genet., 1988, vol. 75, pp. 319-329.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Fesenko, I.A., Khrustaleva, L.I. & Karlov, G.I. Organization of the 378-bp Satellite Repeat in Terminal Heterochromatin of Allium fistulosum. Russian Journal of Genetics 38, 745–753 (2002). https://doi.org/10.1023/A:1016379319030
Issue Date:
DOI: https://doi.org/10.1023/A:1016379319030