Abstract
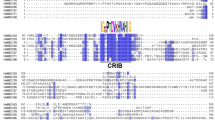
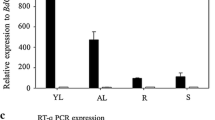
14-3-3 proteins form a family of highly conserved proteins with central roles in many eukaryotic signalling networks. In plants, they bind to and activate the plasma membrane H+-ATPase, creating a binding site for the phytotoxin fusicoccin. Barley 14-3-3 transcripts accumulate in the epidermis upon inoculation with the powdery mildew fungus. We have isolated a cDNA encoding a plasma membrane H+-ATPase (HvHA1), which is also induced by powdery mildew attack. The C-terminal domain of this H+-ATPase interacts with 14-3-3 proteins in the yeast two-hybrid system. Inoculation with the powdery mildew fungus, or treatment with fusicoccin, results in an increase in fusicoccin binding ability of barley leaf membranes. Overlay assays show a fungus-induced increase in binding of digoxygenin-labelled 14-3-3 protein to several proteins including a 100 kDa membrane protein, probably the plasma membrane H+-ATPase. These effects are seen specifically in the inoculated epidermis and not in the whole leaf. We propose that 14-3-3 proteins are involved in an epidermis-specific response to the powdery mildew fungus, possibly via an activation of the plasma membrane H+-ATPase.
Similar content being viewed by others
References
Baunsgaard, L., Fuglsang, A.T., Jahn, T., Korthout, H.A.A.J., de Boer, A.H. and Palmgren, M.G. 1998. The 14–3–3 protein associates with the plasma membrane H+-ATPase to generate a fusicoccin binding complex and a fusicoccin responsive system. Plant J. 13: 661–671.
Booij, P.P., Roberts, M.R., Vogelzang, S.A., Kraayenhof, R. and de Boer, A.H. 1999. 14–3–3 proteins double the number of outwardrectifying K+ channels available for activation in tomato cells. Plant J. 20: 673–683.
Bradford, M.M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein-dye binding. Anal. Biochem. 72: 248–254.
Brandt, J., Thordal-Christensen, H., Vad, K., Gregersen, P.L. and Collinge, D.B. 1992. A pathogen induced gene of barley encodes a protein showing high similarity to a protein kinase regulator. Plant J. 2: 815–820.
Durfee, T., Becherer, K., Chen, P.L., Yeh, S.H., Yang, Y.Z., Kilburn, A.E., Lee, W.H. and Elledge, S.J. 1993. The retinoblastoma protein associates with the protein phosphatase type-1 catalytic subunit. Genes Dev. 7: 555–569.
Felix, G. and Boller, T. 1995. Systemin induces rapid ion fluxes and ethylene biosynthesis in Lycopersicon peruvianum cells. Plant J. 7: 381–389
Finnie, C., Borch, J. and Collinge, D.B. 1999. 14–3–3 proteins: eukaryotic regulatory proteins with many functions. Plant Mol. Biol. 40: 545–554.
Fuglsang, A. T., Visconti, S., Drumm, K., Jahn, T., Stensballe, A., Mattei, B., Jensen, O.N., Aducci, P. and Palmgren, M. G. 1999. Binding of 14–3–3 protein to the plasma membrane H+-ATPase AHA2 involves the three C-terminal residues Tyr946–Thr-Val and requires phosphorylation of Thr947. J. Biol. Chem. 274: 36774–36780.
Gianinazzi-Pearson, V., Arnould, C., Oufattole, M., Arango, M. and Gianinazzi, S. 2000. Differential activation of H+-ATPase genes by an arbuscular mycorrhizal fungus in root cells of transgenic tobacco. Planta 211: 609–613.
Gregersen, P. L., Thordal-Christensen, H., Förster, H. and Collinge, D.B. 1997. Differential gene transcript accumulation in barley leaf epidermis and mesophyll in response to attack by Blumeria graminis f.sp. hordei. Physiol. Mol. Plant Path. 51: 85–97.
Hess, W.R., Golz, R. and Börner, T. 1998. Analysis of randomly selected cDNAs reveals the expression of stress-and defencerelated genes in the barley mutant albostrians. Plant Sci. 133: 191–201.
Jahn, T., Fuglsang, A.T., Olsson, A., Brüntrup, I.M,. Collinge, D.B., Volkmann, D., Sommarin, M., Palmgren, M.G. and Larsson, C. 1997. The 14–3–3 protein interacts directly with the C-terminal region of the plant plasma membrane H+-ATPase. Plant Cell 9: 1805–1814.
Kippert, F. 1995. A rapid permeabilization procedure for accurate quantitative determination of β-galactosidase activity in yeast cells. FEMS Microbiol. Lett. 128: 201–206.
Kølster, P., Munk, L., Stølen, O. and Løhde, J. 1986. Near isogenic barley lines with genes for resistance to powdery mildew. Crop Sci. 26: 903–907.
Larsson, C., Sommarin, M. and Widell, S. 1994. Isolation of highly purified plant plasma membranes and separation of inside-out and right-side-out vesicles. Meth. Enzymol. 228: 451–469.
Marra, M., Olivari, C., Visconti, S., Albumi, C., Aducci, P. and De Michelis, M.I. 2000. A phosphopeptide corresponding to the cytosolic stretch connecting transmembrane segments 8 and 9 of the plasma membrane H+-ATPase binds 14–3–3 proteins and inhibits fusicoccin-induced activation of the H+-ATPase. Plant Biol. 2: 11–16.
Moorhead, G., Douglas, P., Cotelle, V., Harthill, J., Morrice, N., Meek, S., Deiting, U., Stitt, M., Scarabel, M., Aitken, A. and Mackintosh, C. 1999. Phosphorylation-dependent interactions between enzymes of plant metabolism and 14–3–3 proteins. Plant J. 18: 1–12.
Murphy, P.J., Langridge, P. and Smith, S.E. 1997. Cloning plant genes differentially expressed during colonization of roots of Hordeum vulgare by the vesicular-arbuscular mycorrhizal fungus Glomus intraradices. New Phytol. 135: 291–301.
Oecking, C. and Hagemann, K. 1999. Association of 14–3–3 proteins with the C-terminal autoinhibitory domain of the plant plasmamembrane H+-ATPase generates a fusicoccin-binding complex. Planta 207: 480–482.
Oecking, C., Piotrowski, M., Hagemeier, J. and Hagemann, K. 1997. Topology and target interaction of the fusicoccin-binding 14–3–3 homologs of Commelina communis. Plant J. 12: 441–453.
Olsson, A., Svennelid, F., Ek, B., Sommarin, M. and Larsson, C. 1998. A phosphothreonine residue at the C-terminal end of the plasma membrane H+-ATPase is protected by fusicoccininduced 14–3–3 binding. Plant Physiol. 118: 551–555.
Piotrowski, M., Morsomme, P., Boutry, M. and Oecking, C. 1998. Complementation of the Saccharomyces cerevisiae plasma membrane H+-ATPase by a plant H+-ATPase generates a highly abundant fusicoccin binding site. J. Biol. Chem. 273: 30018–30023.
Roberts, M.R. and Bowles, D.J. 1999. Fusicoccin, 14–3–3 proteins, and defence responses in tomato plants. Plant Physiol. 119: 1243–1250.
Schaller, A. and Oecking, C. 1999. Modulation of plasma membrane H+-ATPase activity differentially activates wound and pathogen defense responses in tomato plants. Plant Cell 11: 263–272.
Seehaus, K. and Tenhaken, R. 1998. Cloning of genes by mRNA differential display induced during the hypersensitive reaction of soybean after inoculation with Pseudomonas syringae pv. glycinea. Plant Mol. Biol. 38: 1225–1234.
Sutton, P.N., Henry, M.J. and Hall, J.L. 1999. Glucose, and not sucrose, is transported from wheat to wheat powdery mildew. Planta 208: 426–430.
Thordal-Christensen, H. and Smedegaard-Petersen, V. 1988. Comparison of resistance-inducing abilities of virulent and avirulent races of Erysiphe graminis f.sp. hordei and a race of Erysiphe graminis f.sp. tritici. Plant Path. 37: 20–27.
Thordal-Christensen, H., Brandt, J., Cho, B.H., Gregersen, P.L., Rasmussen, S.K., Smedegaard-Petersen, V. and Collinge, D.B. 1992. cDNA cloning and characterization of two barley peroxidase transcripts induced differentially by the powdery mildew fungus. Physiol. Mol. Plant Path. 40: 395–409.
Thordal-Christensen H., Gregersen, P.L. and Collinge, D.B. 2000. The barley/Blumeria (syn. Erysiphe) graminis interaction: a case study. In: A.J. Slusarenko, R.S.S. Fraser and L.C. van Loon (Eds.) Mechanisms of Resistance to Plant Diseases, Kluwer Academic Publishers, Dordrecht, Netherlands, pp. 77–100.
Thorpe, G.G. and Kricka, L.J. 1986. Enhanced chemiluminescent reactions catalyzed by horseradish peroxidase. Meth. Enzymol. 133: 331–353.
Toroser, D., Athwal, G.S. and Huber S.C. 1998. Site-specific regulatory interaction between spinach leaf sucrose-phosphate synthase and 14–3–3 proteins. FEBS Lett. 435: 110–114.
van den Wijngaard, P.W., Bunney, T.D., Roobeek, I., Schonknecht, G. and de Boer, A.H. 2001. Slow vacuolar channels from barley mesophyll cells are regulated by 14–3–3 proteins. FEBS Lett. 488: 100–104.
van Heusden, G.P., Griffiths, D.J., Ford, J.C., Chin-A-Woeng, T.F., Schrader, P.A., Carr, A.M. and Steensma, H.Y. 1995. The 14–3–3 proteins encoded by the BMH1 and BMH2 genes are essential in the yeast Saccharomyces cerevisiae and can be replaced by a plant homologue. Eur. J. Biochem. 229: 45–53.
Vera-Estrella, R., Barkla, B., Higgins, V.J. and Blumwald, E. 1994. Plant defense response to fungal pathogens: activation of host plasma membrane H+-ATPase by elicitor-induced enzyme dephosphorylation. Plant Physiol. 104: 209–215.
Wei, Y.D., Zhang, Z., Andersen, C.H., Schmelzer, E., Gregersen, P.L., Collinge, D.B., Smedegaard-Petersen, V. and Thordal-Christensen, H. 1998. An epidermis/papilla-specific oxalate oxidase-like protein in the defence response of barley attacked by the powdery mildew fungus. Plant Mol. Biol. 36: 101–112
Wevelsiep, L., Rüpping, E. and Knogge, W. 1993. Stimulation of barley plasmalemma H+-ATPase by phytotoxic peptides from the fungal pathogen Rhynchosporium secalis. Plant Physiol. 101: 297–301.
Xing, T., Higgins, J. and Blumwald, E. 1996. Regulation of plant defence responses to fungal pathogens: two types of protein kinases in the reversible phosphorylation of the host plasma membrane H+-ATPase. Plant Cell 8: 555–564.
Xing, T., Higgins, V.J. and Blumwald, E. 1997. Identification of G proteins mediating fungal elicitor-induced dephosphorylation of host plasma membrane H+-ATPase. J. Exp. Bot. 48: 229–237.
Zhou, F., Andersen, C.H., Burhenne, K., Fischer, P.H., Collinge, D.B. and Thordal-Christensen, H. 2000. Proton extrusion is an essential signalling component in HR of epidermal single cells in the barley-powdery mildew interaction. Plant J. 23: 245–254.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Finnie, C., Andersen, C.H., Borch, J. et al. Do 14-3-3 proteins and plasma membrane H+-ATPases interact in the barley epidermis in response to the barley powdery mildew fungus?. Plant Mol Biol 49, 137–147 (2002). https://doi.org/10.1023/A:1014938417267
Issue Date:
DOI: https://doi.org/10.1023/A:1014938417267