Abstract
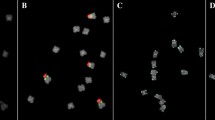
Large variation in genome size as determined by the nuclear DNA content and the mitotic chromosome size among diploid rice species is revealed using flow cytometry and image analyses. Both the total chromosomal length (r_0.939) and the total chromosomal area (r_0.927) correlated well with the nuclear DNA content. Among all the species examined, Oryza australiensis (E genome) and O. brachyantha (F genome), respectively, were the largest and smallest in genome size. O. sativa (A genome) involving all the cultivated species showed the intermediate genome size between them. The distribution patterns of genome-specific repetitive DNA sequences were physically determined using fluorescence in situ hybridization (FISH). O. brachyantha had limited sites of the repetitive DNA sequences specific to the F genome. O. australiensis showed overall amplification of genome-specific DNA sequences throughout the chromosomes. The amplification of the repetitive DNA sequences causes the variation in the chromosome morphology and thus the genome size among diploid species in the genus Oryza.
Similar content being viewed by others
References
Arumuganathan K, Earle ED: Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9: 208–218 (1991).
Aswidinnoor H, Nelson RJ, Dallas JF, McIntyre CL, Leung H, Gustafson JP: Cloning and characterization of repetitive DNA sequences of Oryza minuta and Oryza australiensis. Genome 34: 790–798 (1991).
Bennett MD, Leitch IJ: Nuclear DNA amounts in angiosperms. Ann Bot 76: 113–176 (1995).
Bennett MD, Smith JB: Nuclear DNA amounts in angiosperms. Phil Trans R Soc Lond B274: 227–274 (1976).
Flavell RB: Themolecular characterization and organization of plant chromosomal DNA sequences. Annu Rev Plant Physiol 31: 569–596 (1980).
Fukui K: Plant chromosomes at mitosis. In: Fukui K, Nakayama S(ed) Plant Chromosomes: Laboratory Methods, pp. 1–18. CRC Press, Boca Raton, FL (1996).
Fukui K, Iijima K: Somatic chromosome map of rice by imaging methods, Theor Appl Genet 81: 589–596 (1991).
Fukui K, Kamisugi Y, Sakai F: Physical mapping of 5S rDNA loci by direct-cloned biotinylated probes in barley chromosomes. Genome 37: 105–111 (1994).
Fukui K, Ohmido N, Khush GS: Variability in rDNA loci in the genus Oryza detected through fluorescence in situ hybridization. Theor Appl Genet 87: 893–899 (1994).
Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E: Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220: 1049–1051 (1983).
Gill KS, Gill BS, Endo TR: A chromosome region-specific mapping strategy reveals gene-rich telomeric ends in wheat. Chromosoma 102: 374–381 (1993).
Gupta VS, Gadre SR, Ranjekar PK: Novel DNA sequence organization in rice genome. Biochim Biophys Acta 656: 147– 154 (1981).
Iyengar GAS, Gaddipati JP, Sen SK: Characteristics of nuclear DNA in the genus Oryza. Theor Appl Genet 54: 219–224 (1979).
Katayama T: Radiosensitivity in plants. II. Relationship between radiosensitivity and DNA content per nucleus of diploid species in Oryza. Jpn J Breed 21: 241–246 (1971).
Li XB, Liang CZ, Wu HG, Zhai WX, Huang N, Zhu LH: Isolation and identification of a non-specific tandemly repeated DNA sequences in Oryza species. Theor Appl Genet 92: 702– 708 (1996).
Martinez CP, Arumuganathan K, Kikuchi H, Earle ED: Nuclear DNA content of ten rice species as determined by flow cytometry. Jpn J Genet 69: 513–523 (1994).
McCarthy BJ, Farquhar MN: The rate of change of DNA in evolution. Brookhaven Symp Biol 23: 1–43 (1972).
Moore G, Abbo S, Cheung W, Foote T, Gale M, Koebner R, Leitch A, Leitch I, Money T, Stancombe P, Yano M, Flavell R: Key features of cereal genome organization as revealed by the use of cytosine methylation-sensitive restriction endonucleases. Genomics 15: 472–482 (1993).
Nakajima R, Noma K, Ohtsubo H, Ohtsubo E: Identification and characterization of two tandem repeat sequences (TrsB and TrsC) and a retrotransposon (RIRE1) as genome-general sequences in rice. Genes Genet Syst 71: 373–382 (1996).
Noma K, Nakajima R, Ohtsubo H, Ohtsubo E: RIRE1, a retrotransposon from wild rice Oryza australiensis. Genes Genet Syst (submitted).
Ohmido N, Fukui K: Cytological studies of African cultivated rice, Oryza glaberrima. TheorAppl Genet 91: 212–217 (1995).
Ohmido N, Ohtsubo H, Ohtsubo E, Fukui K: Analysis and utility of chromosome information 80. Variability of the distribution of repeated sequences in several rice species with A genome. Jpn J Breed 45: 47 (1995).
Ohtsubo H, Ohtsubo E: Involvement of transposition in dispersion of tandem repeat sequences (TrsA) in rice genomes. Mol Gen Genet 245: 449–455 (1994).
Price HJ: Nuclear DNA content variation within angiosperm species. Evol Trend Plant 2: 53–60 (1988).
Rees H, Jones GH: Chromosome evolution in Lolium. Heredity 22: 1–18 (1967).
Roupakias DG, Kaltsikes PJ: The effect of telomeric heterochromatin on chromosome pairing of hexaploid triticale. Can J Genet Cytol 19: 543–548 (1977).
SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, Bennetzen JL: Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765– 768 (1996).
Schmidt R, West J, Love K, Lenehan Z, Lister C, Thompson H, Bouchez D, Dean C: Physical map and organization of Arabidopsis thaliana chromosome 4. Science 270: 480–483 (1995).
Suoniemi A, Anamthawat-Jonsson K, Arna T, Schulman H: Retrotransposon BARE-1 is a major, dispersed component of the barley (Hordeum vulgare L.) genome. Plant Mol Biol 30: 1321–1329 (1996).
Vaughan DH: The relationship between the genus Oryza and other grasses. In: The Wild Relatives of Rice, pp. 3–6. International Rice Research Institute, Philippines (1994).
Watanabe Y: Cytogenetic studies on rice and its wild relatives. 1. Cytogenetic investigations of induced polyploids, with special reference to the sterility of synthesized amphiploids. Bull Natl Inst Agric Ser D 26: 1–90 (1975).
Zao X, Wu T, Xie Y, Wu R: Genome-specific repetitive sequences in the genus Oryza. Theor Appl Genet 78: 201– 209 (1989).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Uozu, S., Ikehashi, H., Ohmido, N. et al. Repetitive sequences: cause for variation in genome size and chromosome morphology in the genus Oryza. Plant Mol Biol 35, 791–799 (1997). https://doi.org/10.1023/A:1005823124989
Issue Date:
DOI: https://doi.org/10.1023/A:1005823124989