INTRODUCTION
Q fever is a widespread zoonosis caused by the obligate intracellular bacterium Coxiella burnetii. In humans, 60% of individuals exposed to C. burnetii remain asymptomatic, whereas 40% of patients develop acute Q fever, which generally presents as a mild flu-like but may progress to pneumonia or acute hepatitis with more severe cases requiring hospitalisation [Reference Angelakis and Raoult1]. Many cases of flu-like illness remain undiagnosed and infection is most often diagnosed in the context of outbreaks. Q fever is endemic in the Basque Country (northern Spain), and notification of cases to the System of Microbiological Information (Sistema de Información Microbiológica, SIM) for an epidemiological assessment is compulsory since 2015.
The main reservoirs of C. burnetii are domestic ruminants (cattle, sheep and goats), that can release large amounts of infectious C. burnetii into the environment through milk, urine, faeces and primarily in birth products [Reference Astobiza2–Reference Rodolakis4]. Placenta of infected sheep and goats can contain 109 C. burnetii/gram [Reference Angelakis and Raoult1, Reference Welsh5]. Coxiella is mainly transmitted via inhalation of aerosols containing contaminated dust particles, can persist in the environment for long periods, and has a very low infectious dose [Reference Porter6, Reference Tissot-Dupont7]. However, identifying individual farms as primary source for specific clusters of human cases is a challenge that requires extensive sampling and genotyping samples from animal and human sources. Still, most genotyping methods are cumbersome and require relatively large quantities of DNA [Reference Massung, Cutler and Frangoulidis8].
Here, we describe the clinical and epidemiological investigation of a Q fever outbreak which occurred in January–February 2016 at a work setting with apparently no occupational-associated risk, and how environmental sampling coupled with Real-Time PCR (RTi–PCR) and genotyping allowed the identification of the infection source.
METHODS
Case presentation and epidemiological investigation
On 1 February 2016, a patient was diagnosed with atypical pneumonia at a private clinic. The patient reported similar symptoms among other employees from his working place. Upon being informed, the local Epidemiology Surveillance Unit contacted the company management, who confirmed that five workers were on sick leave due to flu-like illness, including a woman who had been diagnosed with pneumonia in another clinic that weekend. The company, dedicated to the manufacture of hoists and chains, is located in a rural setting in the boundaries between Bizkaia and Araba (Basque Country, northern Spain), with the nearest village being 2 km away. The municipality, that covers an area of 12·43 km2 and has a population ca. 1300, includes the village and little farms scattered around the countryside. The factory occupies a two-storey building, with a manufacturing plant, two small offices and a changing room and bathroom at the ground level, and administration premises, meeting rooms and a common room where workers usually have lunch on the top floor. Ventilation is natural by means of windows and doors. An outdoors marquee, with access for lorries that deliver and collect goods, is used as a store. Behind the main building there is a river, and in front, a road separates the main building from another company.
That same day (1 February), the Occupational Health Authority (OSALAN) was informed. On 5 February, C. burnetii infection was confirmed by Phase II IgM and IgG (1/1280) detection in a patient who was by then hospitalised. An outbreak investigation group was gathered to design an epidemiological investigation and the general practitioner (GP) of the municipality where the factory is located was contacted and requested to collect information regarding patients with clinical symptoms compatible with C. burnetii infection after the middle of January.
Sample collection
Blood samples were collected from the company staff members who agreed to participate in the epidemiological investigation. Environmental samples (dust) were collected at the factory to investigate if workers had been exposed to the pathogen inside the company premises. To search for possible animal sources of infection, the livestock census of the municipality was revised, and blood samples were collected from a representative number of animals in flocks/herds located within a 5 km radius of the human outbreak site. When herds were visited, vaginal swabs were also taken from animals that had given birth recently, to confirm an active infection by C. burnetii.
Laboratory analyses
According to the corresponding area of competence, human samples were analysed at a local Hospital of Bizkaia whereas animal and environmental samples were analysed at NEIKER (Basque Institute for Agricultural Research and Development). Protocols routinely used at each site were performed, which included slight differences. Briefly, presence of IgG antibodies to C. burnetii Phase II in human sera was investigated using a commercially available indirect IFA (immunofluorescence assay) (Vircell SL®, Spain), considering titres ⩾1/256 as positive and a fourfold IgG titre rise in samples collected 3 weeks apart is considered to be diagnostic for recent infection. Phase II IgM antibodies were detected by a commercial indirect ELISA (Vircell SL®, Spain), considering indexes 1·2–2 as weak positive and >2 as positive [Reference Anderson9]. For molecular analyses, 400 µl of human plasma samples were submitted to DNA extraction using MagNA Pure Compact kit and tested by RTi–PCR using a TaqMan probe specific for the transposon-like IS1111 repetitive region of C. burnetii [Reference Denison, Thompson and Massung10]. Samples were analysed in duplicates and RTi–PCR reactions were considered positive when cycle threshold (Ct) values were below 40.
Animal blood samples were collected into tubes without anticoagulant, and after centrifugation sera were tested for the presence of antibodies against C. burnetii using a commercial indirect ELISA test (LSIVET Ruminant Milk/Serum Q Fever kit; Laboratoire Service International, Lissieu, France) as previously reported [Reference Ruiz-Fons11]. Vaginal swabs and dust samples taken on farms and within the company premises were processed for DNA extraction and analysed by RTi–PCR as previously described [Reference Astobiza12, Reference Schets, de Heer and de Roda Husman13]. An internal amplification control (IAC), constructed as previously described [Reference Ros-García14], was included in the assay to monitor for PCR inhibitors; all samples that tested negative for C. burnetii had to be positive to the IAC to exclude inhibition and be considered real negatives.
Samples with a positive RTi–PCR result and a Ct value below 31 were genotyped by multispacer sequence typing (MST) and a 10-loci single-nucleotide polymorphism (SNP) discrimination using RTi–PCR. The procedure for C. burnetii MST genotyping of eight spacers (Cox2, Cox5, Cox18, Cox22, Cox37, Cox51, Cox56 and Cox61) was as previously described [Reference Glazunova15], with small modifications. Briefly, two 4-plex PCR reactions were carried out followed by individual amplifications for each spacer region. Each amplicon was then purified and sequenced. Genotypes were identified by comparison with the database at http://ifr48.timone.univ-mrs.fr/mst/coxiella_burnetii/blast_result.html. Samples were also genotyped using an SNP-based approach that detects 10 discriminatory SNPs by RTi–PCR [Reference Huijsmans16]. Briefly, 10 RTi–PCR reactions were performed per sample, each including two primers and two MGB® TaqMan probes (labelled with VIC and FAM at 5’ end, respectively) to detect point mutations. Each 20 µl PCR mixture contained 625 nM of each primer, 125 nM of each probe, 1 × Taq Mix ABsolute (ThermoScientific) and 5 µl of template DNA. PCR reactions were run on a BioRad platform (CFX96™ RTi–PCR Detection System) using the following program: 15 min at 95°C, and 45 cycles of 3 s at 95°C, and 30 s at 60°C.
Statistical analyses
Parameters included in the questionnaire concerning risk factors of suffering Q fever were analysed by Fisher exact test (categorical variables) or by Mann–Whitney U test (numerical variables) using SPSS Statistic 21. Statistical significance (P < 0·05) was used to reject the null hypothesis of independence between two variables. Attack rates were assessed by Mantel–Haenszel χ 2 test using Epi Info 7.
Ethical considerations
This study did not require ethical approval since outbreaks are routinely investigated according to the Public Health Services’ ethical guidelines to ensure patients safety. Blood samples were obtained by occupational health technicians involved in the study of the outbreak. Written informed consent was obtained from the workers for blood sample collection and personal data collection following legal regulations (Ley Orgánica 15/1995). Data analysis was performed on an anonymised dataset. Animal blood samples were taken solely for the purpose of this study by the veterinary practitioners in charge of the Official Sanitary Campaigns in the Basque Country directed and supervised by the local Animal Health and Welfare Authority (Diputación Foral de Bizkaia & Diputación Foral de Alava) following Spanish ethical guidelines and animal welfare regulations (Real Decreto 53/2013). The collection of this material did not require the approval of the Ethics Committee for Animal Experimentation because they are considered routine veterinary practice. All flock/herd owners had given an informed consent.
RESULTS
Epidemiological investigation
Once the first cases of pneumonia were diagnosed and C. burnetii identified as the causative agent (5 February), an epidemiological investigation was set up to trace the outbreak. All the workers of the affected company and the premises environment, as well as the inhabitants of the municipality who had had clinical symptoms of pneumonia since the middle of January, were included in the study. A confirmed human case was defined as a person who had worked at the company or lived in the municipality between December 2015 and February 2016, and had a laboratory-positive result (specific antibody response – Phase II IgG or IgM), with or without clinical symptoms (fever, pneumonia and/or hepatitis).
On 11 February 2016, the affected company was visited by the outbreak control team integrated by personnel from the Epidemiology Surveillance Unit, the Occupational Health Authority (OSALAN) and Health Prevention Services of the company. Twenty-seven of the 29 workers who had access to the company premises during the outbreak agreed to participate; these included 10 who lived in the village (<2 km of the factory), 13 who lived within 8–25 km of the factory and the remaining four living within a 25–60 km radius. Blood samples collected from the 27 workers were tested for the presence of C. burnetii Phase II IgG and IgM antibodies in sera collected 3 weeks apart (2–15 February and 24 February–3 March). Eight (29·6%) workers (six male, two female) were serology positive; seven showed seroconversion (⩾4-fold increase in IgG antibody titre) and one was IgM positive at first sampling (results from the second sampling were unavailable). Four workers who had IgG positive, yet low stable titres (1/128 or 1/256) and 15 without antibodies, were considered non-case. RTi–PCR analyses performed on plasma were all negative, including the eight confirmed cases. Seven of the confirmed cases presented symptoms such as fever or flu-like signs and five of them had pneumonia; one female (43-years old) who seroconverted was asymptomatic. One of the patients (male, 33-years old) with pneumonia needed to be hospitalised. The attack rate among workers was 29·6%.
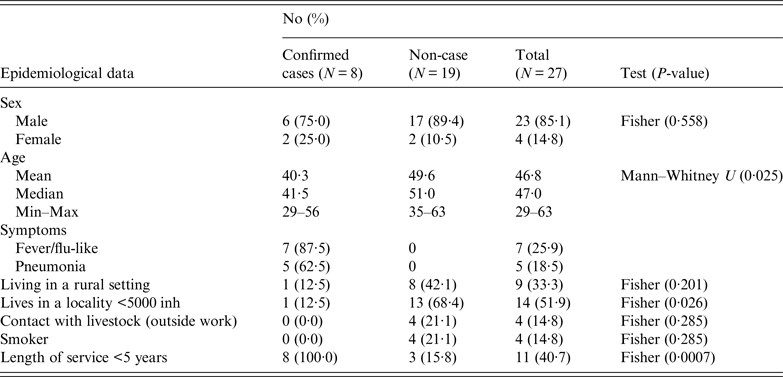
Workers were requested to respond a questionnaire (that included work characteristics, socio-demographical aspects, contacts with livestock and health records) to investigate risk factors of exposure (both at work and elsewhere). The results obtained in the epidemiological questionnaire according to variables and case definition, are summarised in Table 1. The male-to-female ratio of confirmed cases did not differ significantly from the ratio of male-to-female employees (P = 0·558). The four women worked at the offices, three upstairs and one in the ground floor. Most men worked in the manufacturing plant at ground level (17), another five worked in the office downstairs and another one upstairs. However, workers usually moved all over the place, including the marquee. Three of the confirmed cases (male) worked mainly in the manufacturing plant and the other five in the offices (three men at the offices downstairs and two women upstairs). Mean age of confirmed cases was lower than non-affected workers (P = 0·025). None of the affected workers lived in the municipality, although one visited the village regularly during the weekends. Only one of the affected workers lived in a rural area and proportion of cases among those living in a locality with fewer than 5000 inhabitants was significantly lower (P = 0·026). All the workers affected by the outbreak had been working at the company for <5 years (six <1 year). No significant differences in incidence were observed according to smoking habits, living in a rural setting or being in contact with livestock outside work. However, three workers reported contact with domestic ruminants; two, occasionally helped their parents with the cattle they had, and another worker owned a herd of 33 goats located 3 km away from the factory.
Table 1. Summary of the results obtained in the epidemiological questionnaire according to variables and case definition
To investigate any possible cases among the local population, the GP provided serological test results from patients diagnosed with pneumonia or with compatible clinical symptoms after the middle of January. These included 19 local residents, seven of them being senior citizens who lived at the residential home for the elderly. Only one of the 19 patients (a 50-years old woman) was positive to C. burnetti as determined by serology. Laboratory tests performed on blood samples collected from the other patients identified Chlamydia pneumoniae, Streptococcus pneumoniae and Mycoplasma pneumoniae as causes of the pneumonia. Neither clinical symptoms nor cases of sick leave were reported in the company situated in front of the factory affected by the outbreak.
The epidemic curve representing the progression of illnesses onset in confirmed cases (Fig. 1) dates the apparition of first symptoms between January 20 and February 4 among workers, and delays until February 9 when considering other local patients.
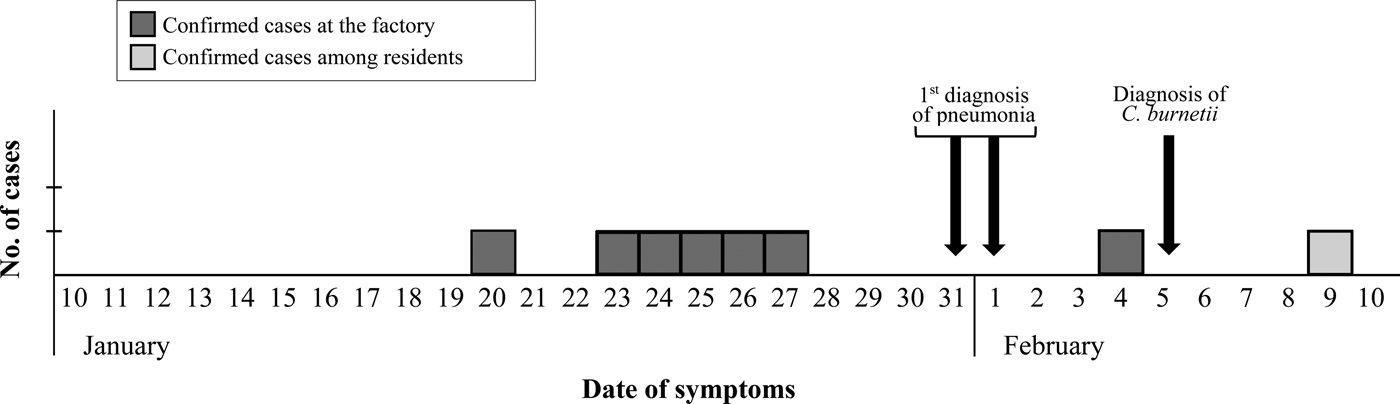
Fig. 1. Epidemic curve based on the onset of symptoms for confirmed cases.
Outbreak source investigation: animal and environmental sampling
ELISA test performed on animal blood samples collected between 24 February and 10 April detected positive animals in two goat flocks, three sheep flocks and one dairy cattle herd (Table 2). Within-herd seroprevalence values were generally low in sheep flocks and in the cattle herd, and vaginal swabs taken from recently lambed ewes in two sheep flocks (30 and 2 swabs, respectively) were all negative. However, in one goat flock, 91·7% (22/24) of the tested animals were seropositive (Table 2). All vaginal swabs (four) and dust samples (four) further collected (21 April) inside the goat farm premises were RTi–PCR positive to C. burnetii (Table 3). The owner of this goat flock turned out to be a worker of the company where the outbreak took place; he did not show any symptoms of disease and did not seroconvert. This flock was suspected to be the source of infection, and the worker considered the likely vehicle for the transmission of the infection.
Table 2. Results on the presence of antibodies against C. burnetii (ELISA test) in animal sera collected from farms within a 5 km radius from the human outbreak
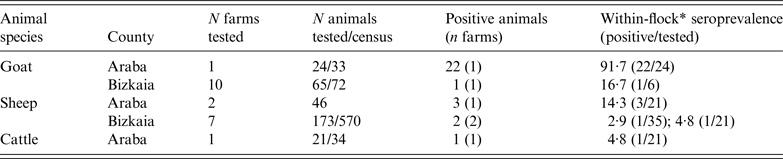
* Seroprevalence within flocks where positive animals were detected (number of positive animals/number of animals tested within the flock/herd).
Table 3. C. burnetii RTi–PCR results on samples collected from the company (workers and dust samples) and the suspected goat farm (animals and environmental dust samples)
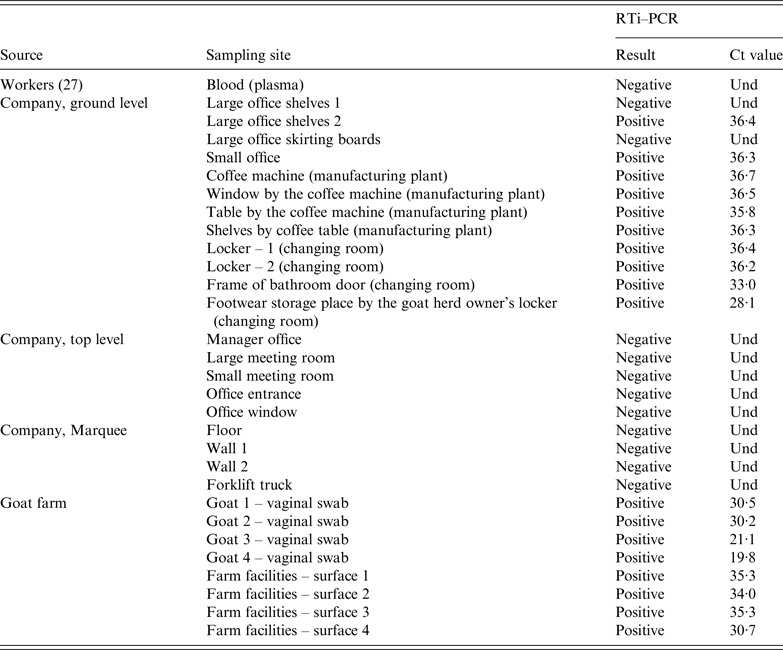
Und, undetermined Ct value since no amplification is produced and sample is therefore identified as negative.
At the factory, 21 dust samples were collected on 3 May from different surfaces throughout the premises in both floors of the building and the marquee. Sampling sites included walls, floors, skirting boards, office shelves, changing room lockers and coffee machine (Table 3). All the samples collected from the top floor (five) were negative. Similarly, samples collected at the marquee (four) were also negative. However, C. burnetii DNA was widespread in the ground floor, with most of the dust samples collected at this level (10/12) still positive 14 weeks after the outbreak. Positive samples concentrated in the office located at the entrance of the manufacturing plant, coffee machine, changing room and the access to the bathroom at the ground level. The sample with the lowest Ct was the one collected by the locker of the goat herd owner (Table 3).
To compare the strains circulating among the animals and at the factory, DNA from RTi–PCR-positive samples (C t < 31) collected at the farm and the company premises were genotyped using MST and a 10 SNP discrimination RTi–PCR assays. The same genotype (MST18 and SNP type 8) was detected in both locations, the farm (four vaginal swabs and one dust sample) and the factory (frame of bathroom door).
DISCUSSION
Q fever is a zoonotic disease that is most commonly associated with outbreaks in work settings that involve contact with animals, such as slaughterhouses workers, veterinarians, farmers, shearers or livestock transport drivers [Reference Angelakis and Raoult1, Reference Bond17, 18]. Here, a C. burnetii outbreak was declared among workers of a company that manufactures hoists and chains, a work setting where animals were not present and therefore with no occupational-associated risk. Although animals were not present in the surroundings of the building, possible animal sources of infection were investigated. On one hand, the questionnaire covered demographic characteristics and exposure categories such as contact with livestock outside work or living in rural areas; on the other, samples were collected from a representative number of ruminants in flocks/herds located within a 5 km radius of the outbreak site. Results from both approaches pointed towards a goat flock with a high percentage of Coxiella-seropositive animals whose owner was an employee of the company where the outbreak took place. To confirm if this goat flock was the source of infection and the worker the likely vehicle for the transmission, environmental samples were collected at the factory and the farm and analysed by molecular methods for strain detection and characterisation.
RTi–PCR results showed the presence of C. burnetii DNA at the farm (both in dust and vaginal swabs), and also at the factory, where dust samples collected from different surfaces of the company facilities were positive. However, positive samples concentrated in the ground floor of the factory, whereas samples collected at the marquee and the top floor were all negative by RTi–PCR. These results suggested that C. burnetii had not entered the premises by the marquee, and that the contamination did not reach the top floor. Distribution of positive samples suggested that contamination had entered through the main entrance reaching intervening zones that needed to be crossed to reach the manufacturing area, such as the little offices by the main entrance, the bathroom and the changing rooms. Remarkably, the sample with the lowest Ct value was the one collected by the locker of the goat herd owner, where he left his footwear. This widespread distribution confirmed the exposure of workers to the infection inside the factory. The most probable vehicle for the bacteria entering the factory was the worker's boots which were worn inside the farm and in the factory. The questionnaire revealed that during the kidding season, the worker visited the animals briefly before leaving for work without changing his footwear, the same boots he wore until reaching the changing room at the company where he changed into working boots. Absence of seroconversion of the owner of the goat flock despite managing the animals himself was unexpected. However, the Basque Country is an endemic region for Q Fever, and in fact, he had IgG positive yet stable titres below 1/256 (and therefore he was considered non-case). This suggests that he had been in contact with C. burnetii before the factory outbreak and, albeit at low levels, antibodies remained in blood protecting him from the disease. Similarly, his family members did not show any symptoms of disease, although they were never tested.
Interestingly, kidding season at the flock took place at the end of December, and first symptoms among workers appeared at the end of January. Considering that incubation of acute Q fever takes 2–4 weeks [Reference Angelakis and Raoult1], exposure was estimated to have extended from the end of December until the end of January. The only case occurred among local residents did not show symptoms until the second week of February, however, no links with the factory or the workers could be established in that case.
This long incubation time might explain why affected workers could only be diagnosed by serology but were all negative by RTi–PCR. More than 3 weeks had probably elapsed since the estimated infection time by the time blood samples were collected. As reported by Schneeberger et al. (2010) [Reference Schneeberger19], the latest time point after onset of disease in which C. burnetii DNA could be detected was at day 17. They concluded that RTi–PCR with serum samples was indispensable for early diagnosis of acute Q fever, but C. burnetii DNA became undetectable as the serological response developed. By the time blood samples were collected in the study herein, most patients already had antibodies.
After the outbreak, a stringent cleaning and disinfection procedure using 1% Virkon® S (Bayer Hispania S.L., Barcelona, Spain) was implemented in the factory, particularly in the areas were infection was detected, and actions were taken to raise farmers’ awareness of the biological risk associated to their job [Reference van den Brom20]. Main points raised included precautions related to management of foetuses, placentas and slurry, as well as biosecurity measures, particularly the use of specific clothes and footwear in animal facilities.
Identifying individual farms as primary source for specific clusters of human cases is a challenge that requires genotyping samples from animal and human sources. Here, environmental sampling coupled with RTi–PCR and genotyping (both by MST and SNP-RTi–PCR) demonstrated that the C. burnetii genotype detected at both sites was the same, thus linking the farm with the factory and identifying the infection source. This study demonstrated once again the importance of environmental sampling in Q fever outbreak investigations to demonstrate that exposition of workers to the pathogen occurred inside the work setting [Reference Alonso21].
Different typing methods have been used to characterize the genetic diversity among C. burnetii isolates [Reference Glazunova15, Reference Huijsmans16, Reference Arricau-Bouvery22, Reference Hornstra23], which sometimes hinders comparison of results. Here, we combined two genotyping methods: MST, a widely used technique [Reference Glazunova15] but rather laborious that requires DNA of high enough quantity and quality for unambiguous typing, which we slightly modified into a ‘nested MST assay’ to circumvent this problem; and, a rapid, sensitive and easy to perform SNP-RTi–PCR that targets a panel of 10 SNPs, seven located in single-copy genes and three located in the multicopy transposon-like IS1111 repetitive region of C. burnetii [Reference Huijsmans16]. Thus, the genotype responsible for the outbreak corresponded to MST18 and SNP type 8. This confirms that SNP type 8 is likely to be (or be closely related to) MST genotype ST18. This correspondence was already suggested [Reference Pearson24] as inferred from results from Tilburg et al. [Reference Tilburg25] and a comparison to the phylogenetic relationships of MST genotypes. Genotype MST18 has been previously found in human and animal (sheep and goats) clinical samples in France, Italy, Romania, Greece, Slovak Republic and Germany [Reference Glazunova15, Reference Tilburg26], and was detected once in a goat placenta in the region where the factory was located, Bizkaia, in 2010 [Reference Astobiza27]. The results obtained in this study demonstrated that C. burnetii genotype MST18/SNP8 originating from goats can cause acute cases of Q fever pneumonia in humans in this part of Europe.
In conclusion, serology was used to diagnose Q fever in the first patients within a few days after the apparition of pneumonia symptoms, which allowed to rapidly designing an epidemiological investigation. Animal investigations pointed towards a goat farm with a high seropositivity rate indicative of an active infection, which happened to be owned and managed by one of the workers of the company where the outbreak occurred. However, identifying individual farms as primary source for specific clusters of human cases in work settings with no occupational-associated risk remains a challenge. Here, it was environmental dust sampling coupled with RTi–PCR and genotyping that enabled ascertaining the source of infection. The same C. burnetii genotype was detected in the goats, the farm environment and dust collected at the factory, thus confirming the infection source.
ACKNOWLEDGEMENTS
The authors would like to thank all the clinicians and laboratory staff involved in the management of the outbreak. The authors also want to acknowledge the collaboration of the staff of the factory during the sampling visit, and the collaboration of Manuela Rodríguez-Vargas (Instituto de Salud Carlos III, Majadahonda, Madrid) in Coxiella genotyping. This study was supported by the Spanish National Institute for Agricultural and Food Research and Technology (INIA RTA 2013-00051-C2-00) and the European Regional Development Fund (ERDF). RAA is the recipient of a predoctoral fellowship from INIA.









