Abstract
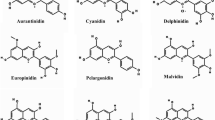
Mechanisms associated with the control of flower color in crape myrtle varieties have yet to be sufficiently elucidated, which has tended to hamper the use of modern molecular and genetic strategies in the breeding programs for this plant. The whole transcriptome of four L. indica varieties characterized by different flower colors (white, light purple, deep purplish pink, and strong red) was sequenced, and we performed bioinformatic, quantitative PCR, and co-expression analyses of R2R3 MYB transcription factor and anthocyanin/flavonol pathway genes. We obtained a total of 49,980 transcripts with full-length coding sequences. Both transcriptome and qPCR analyses revealed that anthocyanin/flavonol pathway genes were differentially expressed among the four different flowers types, with the expression of LiPAL, LiCHS, LiCHI, LiDFR, LiANS/LDOX, and LiUFGT being induced in colorful flowers, whereas that of LiF3´5´H, LiFLS, and LiLAR was found to be inhibited. Base on phylogenetic analysis, seven R2R3 MYB transcriptional factors were identified as putative regulators of flower color. The molecular characteristics and co-expression patterns indicated that these MYBs differentially modulate their target genes, with two probably acting as activators, three as repressors, and one contributing to the regulation of vacuolar pH. The findings of this study indicate that the anthocyanin biosynthesis is more active than the flavonol and proanthocyanin in the colorful flowers. These observations provide new genomic information on L. indica and contribute gene resources for the flower color-targeted breeding of crape myrtle.






Similar content being viewed by others
Data accessibility
The data that support the findings of this study have been deposited into CNGB Sequence Archive (CNSA) of China National GeneBank DataBase (CNGBdb) with accession number CNP0001693.
References
Albert NW, Davies KM, Lewis DH et al (2014) A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26:962–980
Altschul SF, Madden TL, Schaffer AA et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Amato A, Cavallini E, Walker AR et al (2019) The MYB5-driven MBW complex recruits a WRKY factor to enhance the expression of targets involved in vacuolar hyper-acidification and trafficking in grapevine. Plant J 99:1220–1241
Anguraj Vadivel AK, Krysiak K, Tian G, Dhaubhadel S (2018) Genome-wide identification and localization of chalcone synthase family in soybean (Glycine max [L] Merr). BMC Plant Biol 18:325
Apweiler R, Bairoch A, Wu CH et al (2004) UniProt: the universal protein knowledgebase. Nucleic Acids Res 32:D115-119
Ashburner M, Ball CA, Blake JA et al (2000) Gene ontology: tool for the unification of biology. Gene Ontol Consort Nat Genet 25:25–29. https://doi.org/10.1038/75556
Castillejo C, Waurich V, Wagner H et al (2020) Allelic variation of MYB10 is the major force controlling natural variation of skin and flesh color in strawberry (Fragaria spp.) fruit. Plant Cell 32:3723–3749. https://doi.org/10.1105/tpc.20.00474
Chen C, Chen H, Zhang Y et al (2020a) TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13:1194–1202
Chen KX, Wang XM, Zeng HJ, et al (2020b) Cloning and expression analysis of LiANS gene and its correlations with anthocyanin accumulation in Lagerstroemia indica. Zhiwu Fenzi Yuzhong (Molecular Plant Breeding). https://kns.cnki.net/kcms/detail/46.1068.S.20200713.1353.004.html (In Chinese, with English abstract)
Chen M, Xu M, Xiao Y et al (2018) Fine mapping identifying SmFAS encoding an anthocyanidin synthase as a putative candidate gene for flower purple color in Solanum melongena L. Int J Mol Sci 19:789. https://doi.org/10.3390/ijms19030789
Chen Y, Jiang Y, Chen Y et al (2020c) Uncovering candidate genes responsive to salt stress in Salix matsudana (Koidz) by transcriptomic analysis. PLoS ONE 15:e0236129. https://doi.org/10.1371/journal.pone.0236129
Clegg MT, Durbin ML (2000) Flower color variation: a model for the experimental study of evolution. Proc Natl Acad Sci USA 97:7016–7023. https://doi.org/10.1073/pnas.97.13.7016
Deng Y, Li J, Wu S et al (2006) Integrated nr database in protein annotation system and its localization. Comput Eng 32(71–73):76 (In Chinese)
El-Gebali S, Mistry J, Bateman A et al (2019) The Pfam protein families database in 2019. Nucleic Acids Res 47(D1):D427–D432. https://doi.org/10.1093/nar/gky995
Feng X, Zhang Y, Wang H et al (2021) The dihydroflavonol 4-reductase BoDFR1 drives anthocyanin accumulation in pink-leaved ornamental kale. Theor Appl Genet 134:159–169
Finn RD, Bateman A, Clements J et al (2014) Pfam: the protein families database. Nucleic Acids Res 42:D222-230. https://doi.org/10.1093/nar/gkt1223
Fang ZZ, Zhou DR, Ye XF, Jiang CC, Pan SL (2016) Identification of candidate anthocyanin-related genes by transcriptomic analysis of 'Furongli' Plum (Prunus salicina Lindl.) during fruit ripening using RNA-Seq. Front Plant Sci 7:1338. https://doi.org/10.3389/fpls.2016.01338
Fuleki T, Francis FJ (1968) Quantitative methods for anthocyanins. J Food Sci 33:266–274
Han Y, Zhao W, Wang Z, Zhu J, Liu Q (2014) Molecular evolution and sequence divergence of plant chalcone synthase and chalcone synthase-Like genes. Genetica 142:215–225
He D, Liu Y, Cai M et al (2012) Genetic diversity of Lagerstroemia (Lythraceae) species assessed by simple sequence repeat markers. Genet Mol Res 11:3522–3533
Hong Y, Li M, Dai S (2019) Ectopic expression of multiple chrysanthemum (Chrysanthemum × morifolium) R2R3-MYB transcription factor genes regulates anthocyanin accumulation in tobacco. Genes 10:777. https://doi.org/10.3390/genes10100777
Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32:D277-280
Kontturi J, Osama R, Deng X et al (2017) Functional characterization and expression of GASCL1 and GASCL2, two anther-specific chalcone synthase like enzymes from Gerbera hybrida. Phytochem 134:38–45
Koonin EV, Fedorova ND, Jackson JD et al (2004) Comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Bio 5:R7. https://doi.org/10.1186/gb-2004-5-2-r7
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Bio Evol 33:1870–1874
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf 12:323. https://doi.org/10.1186/1471-2105-12-323
Li S, Wang W, Gao J et al (2016) MYB75 phosphorylation by MPK4 is required for light-induced anthocyanin accumulation in Arabidopsis. Plant Cell 28:2866–2883
Lin-Wang K, Bolitho K, Grafton K et al (2010) An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol 10:50. https://doi.org/10.1186/1471-2229-10-50
Liou G, Chiang YC, Wang Y, Weng JK (2018) Mechanistic basis for the evolution of chalcone synthase catalytic cysteine reactivity in land plants. J Biol Chem 293:18601–18612
Luo X, Li H, Wu Z, Yao W et al (2020) The pomegranate (Punica granatum L.) draft genome dissects genetic divergence between soft- and hard-seeded cultivars. Plant Biotechnol J 18:955–968
Ma D, Reichelt M, Yoshida K, Gershenzon J, Constabel CP (2018) Two R2R3-MYB proteins are broad repressors of flavonoid and phenylpropanoid metabolism in poplar. Plant J 96:949–965
Mekapogu M, Vasamsetti BMK, Kwon OK et al (2020) Anthocyanins in Floral colors: biosynthesis and regulation in Chrysanthemum flowers. Int J Mol Sci 21:6537. https://doi.org/10.3390/ijms21186537
Muller J, Szklarczyk D, Julien P et al (2010) eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucleic Acids Res 38:D190-195
Noda N (2018) Recent advances in the research and development of blue flowers. Breeding Sci 68:79–87
Myburg AA, Grattapaglia D, Tuskan GA et al (2014) The genome of Eucalyptus grandis. Nature 510:356–362
Park KI, Nitasaka E, Hoshino A (2018) Anthocyanin mutants of Japanese and common morning glories exhibit normal proanthocyanidin accumulation in seed coats. Plant Biotechnol 35:259–266
Qiao Z, Liu S, Zeng H et al (2019) Exploring the molecular mechanism underlying the stable purple-red leaf phenotype in Lagerstroemia indica cv. Ebony Embers Int J Mol Sci 20:5636. https://doi.org/10.3390/ijms20225636
Qin G, Xu C, Ming R et al (2017) The pomegranate (Punica granatum L.) genome and the genomics of punicalagin biosynthesis. Plant J 91:1108–1128
Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N, Mol J, Koes R (1999) Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cel 11:1433–1444
Robert X, Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320-324
Saigo T, Wang T, Watanabe M, Tohge T (2020) Diversity of anthocyanin and proanthocyanin biosynthesis in land plants. Curr Opin Plant Biol 55:93–99
Seitz C, Ameres S, Schlangen K, Forkmann G, Halbwirth H (2015) Multiple evolution of flavonoid 3’,5’-hydroxylase. Planta 242:561–573
Sharon D, Tilgner H, Grubert F, Snyder M (2013) A single-molecule long-read survey of the human transcriptome. Nat Biotechnol 31:1009–1014
Tatusov RL, Galperin MY, Natale DA, Koonin EV (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36
Wang L, Feng Z, Wang X et al (2010) DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26(1):136–138
Wang XC, Wu J, Guan ML et al (2020) Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant J 101:637–652
Xu H, Zou Q, Yang G et al (2020) MdMYB6 regulates anthocyanin formation in apple both through direct inhibition of the biosynthesis pathway and through substrate removal. Hortic Res 7:72. https://doi.org/10.1038/s41438-020-0294-4
Yahyaa M, Ali S, Davidovich-Rikanati R et al (2017) Characterization of three chalcone synthase-like genes from apple (Malus × domestica Borkh.). Phytochem 140:125–133
Yamagishi M, Shimoyamada Y, Nakatsuka T, Masuda K (2010) Two R2R3-MYB genes, homologs of Petunia AN2, regulate anthocyanin biosyntheses in flower tepals, tepal spots and leaves of asiatic hybrid lily. Plant Cell Environ 51:463–474
Zhang C, Yao X, Ren H, Wang K, Chang J (2019) Isolation and characterization of three chalcone synthase genes in Pecan (Carya illinoinensis). Biomolecules 9:236. https://doi.org/10.3390/biom9060236
Zhang J, Wang LS, Gao JM et al (2008) Determination of anthocyanins and exploration of the relationship between their composition and petal coloration in crape myrtle (Lagerstroemia hybrid). J Integr Plant Biol 50:581–588
Zhang Y, Butelli E, Martin C (2014) Engineering anthocyanin biosynthesis in plants. Curr Opin Plant Biol 19:81–90
Zhao J, Zhang W, Zhao Y et al (2007) SAD2, an importin -like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. Plant Cell 19:3805–3818
Zhou H, Lin-Wang K, Wang F et al (2019) Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation. New Phytol 221:1919–1934
Acknowledgements
The authors are grateful to Yuxing Cao (Nantong Museum), who assisted us in our investigation of L. indica germplasm resources.
Funding
Jian Zhang received funding from the Key Research and Development Program of Jiangsu Province (BE201836); Chunmei Yu received funding from the Jiangsu Province Forestry Science and Technology Innovation and Promotion Project (LYKJ[2019]50) and Modern Agricultural Project of Nantong City (MS12020070); Bolin Lian received the Basic Research Project of Nantong City, China (Grant No. JC2020158); and Wei Fang received funding from the Innovation Program of Undergraduate Students of Jiangsu Province ( 201910304018).
Author information
Authors and Affiliations
Contributions
CY and BL performed transcriptome data analysis and prepared the manuscript; WF, AG, YK, and YJ acquired the data; YC, GL, and FZ participated in the drafting or revision of the manuscript; and JZ conceptualized the research.
Corresponding author
Ethics declarations
Ethical statement
Not applicable.
Conflict of interest
The authors have no competing interests, and all authors have agreed to submit this manuscript to Biologia Futura.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yu, C., Lian, B., Fang, W. et al. Transcriptome-based analysis reveals that the biosynthesis of anthocyanins is more active than that of flavonols and proanthocyanins in the colorful flowers of Lagerstroemia indica. BIOLOGIA FUTURA 72, 473–488 (2021). https://doi.org/10.1007/s42977-021-00094-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42977-021-00094-0