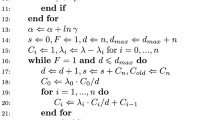Abstract
We study a selective sampling scheme in which survival data are observed during a data collection period if and only if a specific failure event is experienced. Individual units belong to one of a finite number of subpopulations, which may exhibit different survival behaviour, and thus cause heterogeneity. Based on a Poisson process model for individual emergence of population units, we derive a semiparametric likelihood model, in which the birth distribution is modeled nonparametrically and the lifetime distributions parametrically, and define maximum likelihood estimators. We propose a Newton–Raphson-type optimization method to address numerical challenges caused by the high-dimensional parameter space. The finite-sample properties and computational performance of the proposed algorithms are assessed in a simulation study. Personal insolvencies are studied as a special case of double truncation and we fit the semiparametric model to a medium-sized dataset to estimate the mean age at insolvency and the birth distribution of the underlying population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
In survival analysis, missing data are a central notion, as full observability of all units is rarely achievable. Consequently, statistical methods and approaches are designed to consider the various types of missing information to obtain valid inference. The situation we address in the following is at the intersection of survival analysis and retrospective studies, where truncated data result from sampling only failed units within a certain data collection period.
Example 1
Consider personal insolvencies in Germany (see also Sect. 5). When a person becomes insolvent in Germany, individual information on the person is made available due to legal processes. Since the information is accessible for a limited amount of time through a public website, only units with their insolvency event occurring within the data collecting period can be observed. We have collected data from this source from March 2018 to December 2018 and sampled data on insolvent persons born between 1920 and 2000 in all 16 federal states of Germany. Figure 1 illustrates this truncation mechanism.
This sampling design is a special case of random double truncation, as the lifetimes are truncated if they are either too short or too long. Random double truncation may occur as an unavoidable characteristic of data collection procedures or may be chosen deliberately as a cost-effective alternative to other sampling designs. In the example above, the stochastic process of births over time poses a specific component of the truncation setup, which is addressed here.
Random double truncation is relevant to many areas including medicine (see Lagakos et al. 1988; Scheike and Keiding 2006; Moreira et al. 2014), astronomy (see Efron and Petrosian 1999), economics and engineering (see Lawless and Kalbfleisch 1992; Ye and Tang 2016). In recent years, attention to this area of research has been growing (see Emura and Konno 2012; Zhang 2015; Emura et al. 2017; Dörre and Emura 2019; de Uña-Álvarez 2020). This is partly due to the fact that administrative and economically collected data are often observational and frequently subject to selective criteria or constraints which can be posed in a truncation framework.
Among the various existing models and methods for doubly truncated data that have been investigated, the nonparametric maximum likelihood estimator under random double truncation (NPMLE, see Shen 2010a; Moreira and Uña-Álvarez 2010; Emura et al. 2015) has perhaps been studied most extensively. The NPMLE is generally applicable under double truncation, but can be computationally demanding and requires caution regarding identifiability aspects (see Shen 2010a) and the existence of the estimator (see Xiao and Hudgens 2019).
Parametric models have been developed partly to address the drawbacks of the nonparametric approach (see Emura et al. 2017; Dörre and Emura 2019). In Dörre (2020), fully parametric models have been studied in a setting similar to the one studied here. Finally, semi-parametric models (see Wang 1989; Shen 2010b) have been proposed in the context of double truncation.
Most existing models for doubly truncated data are primarily designed for homogeneous populations and can not readily be extended to heterogenous populations in an efficient manner. However, heterogeneity may be an immanent feature of survival data. For instance, in the example described above, it can not be assumed that the underlying lifetime distributions of people in different federal states are identical. While existing approaches such as the NPMLE could be applied to each subpopulation separately, this procedure would not be efficient if the subpopulations have common structural properties.
By incorporating available information on the structure of the study population, we derive a semiparametric method for survival data under double truncation. The survival data are modeled as heterogeneous in that the population may consist of separate subpopulations with potentially different lifetime distributions.
In Sect. 2, we specify the sampling setup and parameter space, and derive a corresponding likelihood model in order to define proper maximum likelihood estimates. By reconsidering the model structure, we address asymptotic properties and discuss notions of point processes and inverse probability weighting in Sect. 3. In Sect. 4, finite-sample properties of the proposed maximum likelihood estimates and numerical optimization methods are examined by means of simulation studies. We apply the methods to study personal insolvencies in Germany in Sect. 5.
2 Model and methods
2.1 General framework
We suppose that the units under investigation experience two subsequent events, generically called ’birth’ and ’failure’, throughout time. The meaning of these events may differ with respect to the particular context, and constitute a survival analysis setup, where the ’lifetime’ is defined as the duration between birth and failure. Furthermore, we suppose that the population of interest is composed of a finite number \(m\) of distinct subpopulations, which may exhibit different lifetime distributions.
Units emerge throughout time according to stochastic processes, yielding random subpopulation sizes \(N_j, j=1,\ldots ,m\). Each unit is born at a random time \(b\) and has a random lifetime \(y\). The corresponding failure event therefore occurs at \(b+ y\). The stochastic process describing the birth of units implicitly corresponds to a distribution \(G(\cdot )\) of births in the population.
Due to the sampling setup, only units that fail within the data collection interval \([\tau ^{\mathrm {L}}, \tau ^{\mathrm {R}}]\) are observed. Therefore, any unit fulfilling the selection criterion
is being sampled, and for sampled units we assume that full information is available, i.e. birth \(b\), lifetime \(y\) and the variable \(x\in \{1,\ldots ,m\}\) indicating the subpopulation to which the unit belongs (see Fig. 2). Units not fulfilling this selection criterion are truncated in the sense that no information about those units is available.
The sample consists of \(n\leqslant N\) units, and is subdivided into random subsamples of sizes \(n_j, j=1, \ldots , m\), according to the considered subpopulations. Note that the subpopulation sizes \(N_j\) are unobservable, whereas the subsample sizes \(n_j\) are observed. We denote the data as \(D_{n}= \{(b_i, x_i, y_i): i =1, ..., n\}\).
Throughout, we make the following assumptions:
-
(A)
For each \(j=1,\ldots ,m\), the units in the jth subpopulation emerge independently of all other subpopulations and stochastically within a common fixed time interval \([s,t]\) according to a Poisson process with intensity function \(b\mapsto \lambda _j(b) > 0, \ b\in [s,t]\).
-
(B)
The intensity functions are proportional, i.e. there is a function \(b\mapsto \lambda _0(b)\) and \(a_j > 0, j= 1, \ldots , m\), such that \(\lambda _j(b) = a_j \lambda _0(b)\).
-
(C)
The lifetime distributions are continuous and parametric, i.e. for each \(j=1,\ldots ,m\), there is a true parameter \(\varvec{\theta }_j\) such that \(f_j(\cdot |\varvec{\theta }_j)\) is the density function of \(y\) conditional on \(x= j\).
-
(D)
The truncation mechanism is such that a unit \((b, x, y)\) from the population is sampled if and only if it fails within \([\tau ^{\mathrm {L}}, \tau ^{\mathrm {R}}]\), i.e. if \(\tau ^{\mathrm {L}}\leqslant b+ y\leqslant \tau ^{\mathrm {R}}\).
-
(E)
The lifetimes \(y\) and births \(b\) are independent.
The notation introduced in Assumption (C) reflects that different subpopulations may possess different distribution classes. Furthermore, we write \(\varvec{\theta }= (\varvec{\theta }_1, \ldots , \varvec{\theta }_{m})\), where needed. Regarding Assumption (B), we generally set \(a_1 = 1\), such that \(\lambda _0(\cdot ) \equiv \lambda _1(\cdot )\), to avoid non-identifiability of the proportionality factors \(a_j, j=2, \ldots ,m\).
Assumption (E) is necessary in some derivations below. In particular, this assumption requires that the lifetime distribution in any subpopulation does not change over the course of time. In specific applications, this assumption may be questionable. For testing the (quasi-)independence assumption, several approaches have been studied; see Tsai (1990) and Martin and Betensky (2005). Recent papers which address the aspect of the independence assumption within the truncation context are Chiou et al. (2018, 2019) and Moreira et al. (2021).
The general framework of the sampling setup. Units emerge in subpopulations according to a Poisson process with proportional intensity functions. Depending on the individual time of birth \(b\) and lifetime \(y\), a unit is being sampled if it fulfills the selection criterion \(\tau ^{\mathrm {L}}\leqslant b+ y\leqslant \tau ^{\mathrm {R}}\)
2.2 Proportional birth intensity model
The distribution function of a randomly chosen birth \(b\) generated by a Poisson process with intensity function \(\lambda (\cdot )\) is equal to
By Assumption (B) and Eq. (2), it follows that there is a common birth distribution for all considered subpopulations, such that the birth distribution functions of the subpopulations fulfill \(G_j \equiv G\) for all \(j=1, \ldots , m\).
By Assumptions (A) and (D), the observed units of any subpopulation j form a random point process with regard to the observed births \(b_i\) where \(x_i = j\). It can be shown that the observed point process of each subpopulation j is also a Poisson process. Furthermore, the sum of all observed birth processes is a Poisson process, too, by the independence stated in Assumption (A). For any subpopulation \(j=1,\ldots ,m\), we denote the cumulative intensity of the latent birth process as \(\varLambda _j := \int _{s}^{t} \lambda _j(b) \, \mathrm{d}b^{}\) and the cumulative intensity of the observed birth process as \(\varLambda ^{\mathrm {obs}}_j := \int _{s}^{t} \int _{\mathbb {R}}^{} \lambda _j(b) f_j(y|\varvec{\theta }_j)\mathrm {I}(\tau ^{\mathrm {L}}\leqslant b+ y\leqslant \tau ^{\mathrm {R}}) \, \mathrm{d}y^{} \, \mathrm{d}b^{}\).
The probability of observing one unit, conditional on it belonging to subpopulation j with given birth time \(b\), denoted as \(\pi _j(\varvec{\theta }_j|b)\), depends on the lifetime distribution of the corresponding subpopulation, and is equal to
The intensity function of the observed point process of subpopulation j is equal to
Similarly, \(\varLambda ^{\mathrm {obs}}_j = \varLambda _j \overline{\pi }_j(\varvec{\theta }_j|G)\), where
is the probability of observing a random unit from subpopulation j, given birth distribution function G. To see this, note that
Using Eq. (2), it follows that \(\lambda _j(b) \mathrm{d}b= \varLambda _j \mathrm{d}G(b)\), and therefore
By \(h(\cdot )\), we denote the density function of \(x\), i.e. the probability of a unit from the population belonging to a specific subpopulation. It is a consequence of Assumption (B) that this density function only depends on \({\varvec{a}}= (a_1, \ldots , a_{m})\), representing a multinomial experiment with
Furthermore, the sum of all latent birth processes itself forms a Poisson process, and has cumulative intensity \(\varLambda _1 \sum _{j=1}^m a_j\). Similarly, it holds that the sum of all observed birth processes is a Poisson process, too, for which the cumulative intensity is equal to \(\varLambda _1 \sum _{j=1}^m a_j \overline{\pi }_j(\varvec{\theta }_j|G)\). The probability of observing a single random unit from the population is equal to
where \(\overline{\pi }_j(\varvec{\theta }_j|G)\) is defined as in Eq. (4). Finally, we state the following result, which is fundamental for constructing the likelihood function.
Lemma 1
Under Assumptions (A)–(E), the point process \(\eta (b, x, y) := \sum _{i=1}^{n} \mathrm {I}(b_i \leqslant b, x_i \leqslant x, y_i \leqslant y)\), which counts the observed units in the three-dimensional space \([s,t]\times \{1, \ldots , m\} \times \mathbb {R}_{\geqslant 0}\), is a Poisson process with intensity function
The cumulative intensity is equal to \(\pi (G, {\varvec{a}}, \varvec{\theta }) \varLambda _1 \sum _{j=1}^{m} a_j\).
Proof
It can be shown by elementary calculations that \(\eta (\cdot )\) is Poisson distributed with the corresponding intensity. Assumption (A) ensures independent increments of \(\eta (\cdot )\). A detailed proof is omitted. \(\square \)
2.3 Likelihood function
Under Assumptions (A)–(E), the observed Poisson process has the density function (see Snyder and Miller 1991 and Theorem 3.1.1 in Reiss 1993):
Note that Eq. (7) does not consist of iid factors and is therefore not ideal for likelihood inference. By conditioning on the random sample size \(n\), however, we construct a likelihood function with proper iid terms. For more technical details in a similar model, see Dörre (2020).
To estimate the birth distribution function G nonparametrically, we place positive probability weights precisely on all observed births \(b_1, \ldots , b_n\). Specifically, we set \(\varDelta G_i = \mathrm {P}(b= b_i)\) for each \(i=1,\ldots ,n\), and treat \(\varDelta G_i\) as parameters of the unknown birth distribution. The corresponding birth distribution results as \(G(b) = \sum _{i=1}^{n} \varDelta G_i \cdot \mathrm {I}(b_i \leqslant b)\) for \(b\in [s,t]\). By conditioning Eq. (7) on the random sample size \(n\), we obtain the corresponding likelihood function
where \(\varDelta G= (\varDelta G_1, \ldots , \varDelta G_{n}) \in [0,1]^{n}\) with \(\sum _{i=1}^{n} \varDelta G_i = 1\), \({\varvec{a}}= (a_1, \ldots , a_{m}) \in \mathbb {R}_{> 0}^{m}\) with \(a_1 = 1\) and \(\varvec{\theta }= (\varvec{\theta }_1, \ldots , \varvec{\theta }_{m}) \in \varTheta _1 \times \cdots \times \varTheta _{m}\). We define the maximum likelihood estimator \((\widehat{G}^{(n)}, \varvec{\widehat{a}}, \varvec{\widehat{\varvec{\theta }}})\) as the maximizer of Eq. (8), i.e.
Altogether, this model entails \(n+ m+ \sum _{j=1}^{m} \dim (\varTheta _j) - 2\) parameters, not counting \(a_1\) and \(\varDelta G_{n}\) due to being subject to constraints. The model is semiparametric in the sense that the number of parameters increases linearly with growing sample size. The likelihood function Eq. (8) can be rearranged as
so that the log-likelihood function can be written as
where \(\pi (G, {\varvec{a}}, \varvec{\theta })\) is obtained as in Eq. (6) and, following Eq. (4),
for any distribution G with discrete jumps \(\varDelta G_i\) at \(b_1, \ldots , b_n\).
Note that the likelihood function Eq. (8) represents an inverse probability weighting (IPW) approach, where the probability weights \(\pi (G, {\varvec{a}}, \varvec{\theta })\) are determined endogenously and equally for every observed unit. By conditioning on certain parts of the data, other inverse probability weighting likelihood functions are conceivable under the considered truncation scheme. For instance, for any \(j = 1, \ldots , m\), an estimator \(\widehat{\varvec{\theta }}{}_j^c\) of \(\varvec{\theta }_j\) may be defined as the maximizer of
and \({\widetilde{L}}_j\) is valid in the sense that \(\widehat{\varvec{\theta }}{}_j^c\rightarrow \varvec{\theta }_j\) as \(n_j \rightarrow \infty \), which is not proved here. Using Eq. (8) has the advantage of leading to generally more precise estimates due to the stronger assumptions with regard to the birth distribution, while Eq. (9) has the advantage of not being vulnerable to misspecification with regard to the lifetime distribution in other subpopulations than j or the birth distribution G.
2.4 Birth intensity estimation
The proposed likelihood model Eq. (8) can be regarded as an inverse probability weighted approach, in which every sampled unit is weighted by the inverse probability of observing a single random unit from the population. Roughly speaking, every sampled unit corresponds to \(\pi (G, {\varvec{a}}, \varvec{\theta })^{-1}\) hypothetical units in the population, out of which one single unit fulfills the selection criterion in Eq. (1) and has thus been sampled. Similarly, a sampled unit from the jth subpopulation with birth \(b_i\) corresponds to \(\pi _j(\varvec{\theta }_j|b_i)^{-1}\) hypothetical units before the selection. Based on this perspective and following Eq. (3), we define
as an estimate of the cumulative birth intensity of the jth subpopulation.
The IPW perspective also offers an alternative approach to estimate the proportionality factors \(a_2, \ldots , a_{m}\). Due to the proportional intensity assumption, it holds that \(\varLambda _j = a_j \varLambda _1\) for \(j=2, \ldots , m\). One simple way to estimate the coefficients \(a_2, \ldots , a_{m}\) is therefore to use Eq. (10), by which
is obtained. Note also that this approach is feasible whenever a consistent estimate of \(\varvec{\theta }\) is available, and no estimate of G is required. We show in Sect. 4, that this method leads to valid, yet inefficient estimates.
3 Large-sample properties
Studying the asymptotic behaviour of the proposed estimators is based on the subsample sizes growing indefinitely. We note, however, that \(n_j \rightarrow \infty \) if and only if \(\varLambda _j \rightarrow \infty \), and thus asymptotics can not be studied without changing the birth intensity function. As a consequence of this perspective, it is not possible to estimate \(\varLambda _j\) consistently in the sense that \(|\widehat{\varLambda }_j - \varLambda _j|\) converges to 0 for \(n\rightarrow \infty \). Nevertheless, Eq. (2) shows that the birth distribution G, which we estimate, is invariant with respect to scaling the birth intensity function \(\lambda \), as long as its shape is not changed.
Due to Assumptions (A)–(E), \(n_j\) is Poisson distributed with mean \(\varLambda ^{\mathrm {obs}}_j = \varLambda _j \overline{\pi }_j(\varvec{\theta }_j|G)\). Therefore, when \(\varLambda _j \rightarrow \infty \), it holds that \(n_j \rightarrow \infty \), almost surely, and the expression
converges to \(E(\pi _j(\varvec{\theta }_j|b)^{-1}|x= j)\) by the law of large numbers, where \(E(\cdot )\) is the conditional expectation with respect to the law of observed units in subpopulation j. It holds that
where
is the density function of observed random births \(b\) in subpopulation j. Therefore,
and thus
since \(n_j\) is Poisson distributed with parameter \(\varLambda ^{\mathrm {obs}}_j\). This derivation shows that \(\widehat{\varLambda }_j\) as defined in Eq. (10) is a reasonable estimate of \(\varLambda _j\). Consequently, \(\widehat{a}_j^{\mathrm {simple}}\) as defined in Eq. (11) is expected to be a reasonable estimate of \(a_j\).
To study the large-sample properties of the proposed estimators based on Eq. (8), we consider the asymptotic behaviour of the log-likelihood function. Define
as the stochastic criterion function, which is maximized by the maximum likelihood estimator \((\widehat{G}^{(n)}, \varvec{\widehat{a}}, \varvec{\widehat{\varvec{\theta }}})\). Suppose first that the true birth distribution G is known. A conditional criterion function can be defined as
and it holds due to the law of large numbers that
almost surely, where \(E_{{\varvec{a}}, \varvec{\theta }}\) denotes the expectation with respect to the distribution conditional on \(\tau ^{\mathrm {L}}\leqslant b+ y\leqslant \tau ^{\mathrm {R}}\). For known G, the density function of an observed random pair \((x, y)\) under truncation is precisely \(f_{x}(y|\varvec{\theta }_{x})h(x|{\varvec{a}})/\pi (G, {\varvec{a}}, \varvec{\theta })\), such that Eq. (12) in fact represents the Kullback–Leibler divergence with respect to \(({\varvec{a}}, \varvec{\theta })\) up to a constant additional term. It is well-known that this expression is uniquely maximized at the true parameter values \(({\varvec{a}}, \varvec{\theta })\) under appropriate regularity assumptions (see Lemma 5.35 in van der Vaart 1998).
Now, more generally, assume that some consistent estimator of G is available, i.e. an estimator \(\widehat{G}^{(n)}\) based on the data \(D_{n}\) such that
almost surely, for \(n\rightarrow \infty \). Reconsidering Eq. (4) and Eq. (6) and \(0 < \overline{\pi }_j(\varvec{\theta }_j|G)\leqslant 1\) for every \(j=1,\ldots ,m\), it follows that
Therefore, \(|Q_{n}({\varvec{a}}, \varvec{\theta }|\widehat{G}^{(n)}) - Q_{n}({\varvec{a}}, \varvec{\theta }|G)| \rightarrow 0\) for any \({\varvec{a}}\) and \(\varvec{\theta }\) and thus
as in Eq. (12). It follows that \(\varvec{\widehat{a}}\) and \(\varvec{\widehat{\varvec{\theta }}}\) are consistent under appropriate regularity assumptions, if \(\widehat{G}^{(n)}\) is a consistent estimator of G.
Assuming asymptotic normality of \(\varvec{\widehat{\gamma }}= (\varvec{\widehat{a}}, \varvec{\widehat{\varvec{\theta }}})\) with the variance given by the Fisher information, it is possible to derive asymptotic confidence intervals in a conventional way. To do so, we abbreviate \(\varvec{\gamma }= ({\varvec{a}}, \varvec{\theta })\) and define the matrix \(A_{n}\) of all negative partial derivatives of the log-likelihood function
Given that \(A_{n}\) is a regular matrix, the asymptotic variance matrix of \(\varvec{\widehat{\gamma }}\) can be estimated by the inverse \(\widehat{V}_{n} := A_{n}^{-1}\). The level \((1-\alpha )\) confidence interval for the jth component of \(\varvec{\gamma }\) is then obtained as
where \(z_{1-\alpha /2}\) is the \((1-\alpha /2)\) quantile of the standard normal distribution.
Similarly, confidence intervals for aspects of the latent distribution, say the mean lifetime \(E(y|\varvec{\theta }_j)\) in the jth subpopulation, can be defined. To do so, we first generate a large number M of iid draws
An estimated level \((1-\alpha )\) confidence interval of the mean lifetime is then obtained using the \(\alpha /2\) and \(1-\alpha /2\) quantiles of \(\{E(y|\varvec{\theta }_j^1), \ldots , E(y|\varvec{\theta }_j^M)\}\).
4 Numerical implementation and simulation
4.1 Numerical implementation
To find the maximum likelihood estimate for given data \(D_{n}\), we propose a Newton–Raphson-type algorithm. In this procedure, the stopping criterion is defined with respect to a distance function which indicates the average change in an iteration with respect to the different parameter types. We define the distance function
in which the number \(n\) of support points is incorporated into measuring the distance of two distribution functions \({\widetilde{G}}\) and G. The following algorithm is proposed for determining the maximum likelihood estimate.

For the initial values \(\varvec{\theta }_j^0\), we use the maximum likelihood estimates of \(\varvec{\theta }_j\) in the model without truncation. The key step in this algorithm is that the initial value \(G^0\) is determined utilizing the initial values \(\varvec{\theta }_j^0\) and using Eq. (10). Intuitively, for every i, \(N_i^0\) is the hypothetical number of population units among which the single unit i has been observed. By standardizing these hypothetical population numbers as done in Step 4, the corresponding distribution function is derived. It turns out (see Sect. 4.2), that choosing \(\varDelta G^0\) in this way remarkably reduces the computational runtime in comparison to alternative optimization strategies.
The damping factor \(0 < \omega \leqslant 1\) is used to avoid obtaining invalid parameter values, such as \(\varDelta G_i \leqslant 0\) or \(a_j\leqslant 0\) for any \(j \in \{1, \ldots , m\}\). Depending on the sample size, \(\omega \) may need to be chosen close to zero initially. When \(\varvec{\gamma }^\nu \) is sufficiently close to the maximum likelihood estimator, the damping factor can be dropped, i.e. \(\omega = 1\). Similarly, the Newton–Raphson method in Step 3 of the algorithm MNS may potentially yield inadmissible values \(a_j\) due to numerical reasons. To avoid this issue, one may need to adjust the optimization to ensure robustness (e.g., using a damping factor).
4.2 Simulation studies
4.2.1 Simulation study on validity
We formulate the following simulation setup to study the finite-sample properties of the proposed method (see Table 1). Population units are born in the time interval \([s, t] = [0, 8]\). In all considered setups, we consider \(m=2\) subpopulations and set \(a_2 = 1.5\). The birth intensity function of subpopulation 1 is defined as a piecewise constant function \(\lambda _1(b) = \lambda _{11} \mathrm {I}(b\leqslant (t+s)/2) + \lambda _{12} \mathrm {I}(b> (t+s)/2)\). Lifetimes are exponentially distributed according to \(y\mapsto f_j(y|\theta _j) = \theta _j\exp (-\theta _jy)\) with parameters \(\theta _1\) and \(\theta _2\) in the respective subpopulations. For each simulation setup, we generate 1000 random datasets and determine the estimates using the algorithm MNS. The damping factor is set as \(\omega = 1\) and \(\epsilon = 10^{-4}\).
Following Table 1, comparisons between the Setups 1–2 (\(\lambda _{11} > \lambda _{12}\)) and Setups 3–4 (\(\lambda _{11} = \lambda _{12}\)) are of primary interest, as these setups respectively concern the same birth distributions. The Setups 5 and 6 differ from Setup 4 only in that \(\tau ^{\mathrm {R}}\) is increased. The probability of observing a random unit ranges from 0.138 to 0.261 in the considered setups. Note also that although the birth distribution is in fact parametric in these simulation setups, the birth distribution is estimated nonparametrically.
The simulation results (see Table 2) indicate that the proposed method yields valid estimates of the lifetime parameters \(\varvec{\theta }\). While the bias of the estimates is notable due to the relatively small sample sizes, the MSEs are adequately small and decrease with growing sample size (see Setup 1 vs. 2 and Setup 3 vs. 4). Comparing the Setups 5 and 6 with Setup 4, the MSE is not substantially decreased despite the increased sample size (and increased selection probability). This shows that the structure of the truncation mechanism, including the definition of the data collection period, may influence the precision of estimates in a nontrivial manner, and larger samples not necessarily correspond to higher precision.
Table 3 summarises the performance of \(\widehat{a}_2\) and the comparison to the simple estimate \(\widehat{a}^{\mathrm {simple}}_2\). It turns out that the biases of both estimates are considerable, yet of a similar magnitude. While \(\widehat{a}^{\mathrm {simple}}_2\) is a valid estimator of \(a_2\), \(\widehat{a}_2\) is more precise in all considered setups.
Finally, we study the performance of the algorithm MNS and the estimated birth distribution. For the latter purpose, we define the distances
The results displayed in Table 4 demonstrate that the initial values as defined in Step 2 in the algorithm MNS are suitable for optimization. On average, the first Newton–Raphson-type search in Step 3 in the algorithm MNS required about 6 to 8 iterations before reaching the stopping criterion. Considering that the number of parameters in the studied simulation setups is \(n+3\), requiring on average less than 40 iterations in the final Newton–Raphson-type search in Step 5 in the algorithm MNS seems quite satisfactory.
Assessing the estimation of the birth distribution G, the mean distance of \(\widehat{G}^{(n)}\) to G, denoted as \(E(\Vert G^0 - G\Vert _{\infty })\) is remarkably small in all considered setups and generally decreases with growing sample size, ceteris paribus. Note also that the distance of \(G^0\) to \(\widehat{G}^{(n)}\), denoted as \(E(\Vert G^0 - \widehat{G}^{(n)}\Vert _{\infty })\), is relatively small, again demonstrating that the suggested choice of initial values is appropriate. Nevertheless, \(\Vert G^0 - G\Vert _{\infty } \leqslant \Vert G^0 - \widehat{G}^{(n)}\Vert _{\infty } + \Vert \widehat{G}^{(n)}- G\Vert _{\infty }\), and the final Newton–Raphson method in Step 5 in the algorithm MNS with regard to all parameters is necessary to find the maximum likelihood estimate.
4.2.2 Simulation study on computational performance
We further evaluate the proposed initial parameter choice in the Newton–Raphson algorithm by comparing its performance to reasonable alternatives. A local random search (LRS) is a conceptually simple approach to finding local maxima of the likelihood function (see Zhigljavsky 1991). Given numerically permissible initial values, the algorithm successively finds parameters with higher likelihood value, until a defined termination criterion is met. Reasonable termination criteria are, e.g., that no improvements are found for a defined number of steps or that a preset number of iterations has been performed. The algorithm LRS has the advantages of using computationally inexpensive iterations and of being relatively robust in that the local step sizes can be chosen sufficiently conservative to avoid numerically non-permissible parameters. The algorithm LRS does not utilize the local shape of the log-likelihood function, but just requires the function’s value at candidate points. See the Appendix for an implementation of the algorithm LRS.
In addition, we consider a Newton–Raphson-type algorithm, which we call ’Naive Newton–Raphson Search’ (NNS, see Appendix). This algorithm addresses a drawback of the algorithm LRS using the local shape of the log-likelihood function for updating parameter values in each iteration. The algorithm NNS differs from the algorithm MNS in that the initial birth distribution is chosen naively, i.e. without utilizing the IPW relation expressed in Eq. (10).
We apply the three algorithms on 100 simulated datasets each with the following three setups. Births are randomly drawn in the interval \([s,t] = [0,8]\) from \(m=2\) subpopulations with intensity \(\lambda _1(\cdot ) \equiv \lambda _1 \in \{100, 200, 500\}\) and proportion factor \(a_2 = 1\). Lifetimes are exponentially distributed according to \(f_j(y|\theta _j)= \theta _j\exp (-\theta _jy)\) with parameters \(\theta _1 = 0.4\) and \(\theta _2 = 0.15\). If a unit fails within \([\tau ^{\mathrm {L}}, \tau ^{\mathrm {R}}] = [8, 10]\), it is sampled.
For each algorithm and iteration, the distance of the current log-likelihood value to the maximal value is calculated. For the algorithms NNS and the MNS, 10 iterations have been performed, and took roughly the same runtime. Since an iteration in the algorithm LRS is computationally much cheaper than for the algorithms NNS and MNS, the number of iterations of the algorithm LRS has been adjusted to account for this difference. Therefore, the runtime of all three algorithms has been approximately the same in each setup. The initial values are chosen as \(\theta _j^0 = n_j/\sum _{i: x_i = j} y_i\) for each \(j \in \{1, 2\}\), which would be the subpopulation-specific maximum likelihood estimate of \(\theta _j\) if the data were not subject to truncation, and \(a_2^0 = 1\).
Distance to the maximum log-likelihood value for the algorithms NNS, LRS and MNS. (top panel: \(\lambda _1 = 100\), middle panel: \(\lambda _1 = 200\), bottom panel: \(\lambda _1 = 500\); dashed lines indicate the 80% pointwise confidence band; *The number of iterations of the algorithm LRS have been adjusted to yield comparable runtime to the algorithms NNS and MNS)
Figure 3 shows that the three considered algorithms seem to form a clear order with regard to performance. Due to having the same initial values, the algorithms LRS and NNS start with the same log-likelihood value. The algorithm LRS quickly improves over the algorithm NNS, and then progresses rather slowly towards the maximum. The algorithm MNS clearly has the best performance of the three considered algorithms in the considered setups. Interestingly, the initial value of the algorithm MNS has higher log-likelihood value than the algorithms NNS or LRS achieve within the considered runtime. Moreover, after few steps, the algorithm MNS essentially reaches the maximum of the log-likelihood. This simulation demonstrates that a model-driven initial choice of the parameters \(\varDelta G_i\) can remarkably reduce the runtime of the numerical optimization.
5 Application
Personal insolvency occurs when a person is unable to pay their debts to other entities. In Germany, personal insolvencies are reported publicly due to legal regulations. Reports on the individual insolvencies are available through a public website for 14 days, before public access is removed again. The sample, which results from continually tracking such public reports, includes information on all persons who become insolvent during the timespan of data collection, and is thus subject to the described double truncation framework. From March 2018 to December 2018, \(n= 73{,}370\) units that were born between 1920 and 2000 have been sampled (see Table 5 for basic summary statistics).
Individual information includes the date of birth \(b_i\), the age at insolvency \(y_i\) (adjusted to biological age 18, when legal age is reached in Germany) and the federal state \(x_i\) in which the insolvency is reported. There are \(m=16\) federal states, and they constitute the subpopulations in this analysis. For the lifetimes in any subpopulation \(j \in \{1, \ldots , 16\}\), we assume a Gamma distribution \(\mathrm {Ga}(\alpha _j, \beta _j)\), defined as
In this notation, \(\varvec{\theta }_j = (\alpha _j, \beta _j)\) and \(\varvec{\theta }= ((\alpha _1, \beta _1), \ldots , (\alpha _{16}, \beta _{16}))\). The mean lifetime in the jth subpopulation is equal to \(\alpha _j/\beta _j\). As stated in the Assumptions (A)–(E), we implicitly assume that births are formed independently across the \(m=16\) federal states, and that the corresponding intensity functions are proportional. We have considered this assumption by checking the observed empirical birth distributions for each state in the dataset, and while the impact of the lifetime distributions is neglected via this inspection, we found that there is no apparent reason to discard the proportionality assumption. Note that Assumption (D) is fulfilled by the procedure the data have been collected. Since there is perhaps a substantial percentage of people who never experience insolvency (i.e. ’immortal’ units), the population of interest is implicitly defined as the population of all people in Germany who eventually experience insolvency. Therefore, the birth distribution estimated under the truncation setup does not necessarily represent the actual birth distribution known for Germany. The model altogether contains 73,417 parameters. We apply the algorithm MNS to determine the maximum likelihood estimates.
As initial values in Step 1 of the algorithm MNS, we choose \(a_j^0 = n_j/n_1\). Reasonable initial values \((\alpha _j^0, \beta _j^0)\) were obtained by first fitting the model to a random subsample having size 2000. We choose \(\omega = 0.4\) as the damping factor. The resulting estimates are displayed in Table 6.
Estimates of the coefficients \(a_j\) and estimated mean biological ages at insolvency for \(m=16\) federal states in Germany. Squares left to the federal state names indicate the estimated proportion factors \(\widehat{a}_j\), where darker squares indicate higher birth intensity. Numbers in parentheses indicate the mean ages at insolvency. Vertical bars display the 90% confidence interval of the mean lifetime for each subpopulation
Figure 4 summarizes the estimates of the proportion factors and the mean age at insolvency for each subpopulation. The results indicate distinct differences between the federal states, with the estimated proportion factors generally reflecting the proportions of the federal state sizes known for Germany (numerical values not displayed).
Similarly, the estimated mean age at insolvency varies across federal states, ranging from 56.4 in Saxony-Anhalt to 70.6 in Bavaria. In comparison to the observed mean ages at insolvency (see Table 5) in the sample, the estimates are considerably larger. Without accounting for the truncation mechanism, the mean ages are systematically underestimated. We note that larger subsample sizes correlate with smaller confidence intervals for the mean biological age at insolvency in the respective subpopulations, which is not unexpected.
Figure 5 indicates that the truncation mechanism substantially distorts the birth distribution, as the empirical distribution function differs significantly from the estimate. In particular, more recent births between 1985 and 2000 seem to be overrepresented in the sample in comparison to the population of interest, while people born between 1940 and 1980 are underrepresented.
6 Discussion
We have focused on a joint model for the birth distribution and the lifetime distribution for heterogeneous survival data under double truncation. In doing so, we have derived maximum likelihood estimates for all involved parameters.
The situation is cast as a high-dimensional problem, where the number of parameters increases linearly with sample size. The main advantage is that the model can be treated with well-known concepts for parametric models, and implemented in a standard fashion. One substantial drawback of the large number of parameters is that the optimization method for finding the maximum likelihood estimates needs to be carefully adjusted.
Concerning the implementation of the model, we have demonstrated that caution in the choice of initial values in a Newton–Raphson-type algorithm is important. Using a generic choice is not ideal and can lead to impractical runtime. Instead, we propose model-driven initial values, which greatly reduce computational runtime. In our calculations, we have found that with this approach, the model can be fitted to even medium-sized datasets such as considered in the application (\(n> 50{,}000, \ m>10\)) within reasonable runtime. However, we note that further improvements on the optimization method are certainly possible and worth studying. This also concerns the choice of the damping factor \(\omega \), which has been introduced to avoid obtaining invalid parameter values within the optimization algorithm. We have not included a general guideline on how to properly choose \(\omega \), because it may depend on many variables such as the sample size, the setting of the birth interval [s, t], the data collection period \([\tau ^L, \tau ^R]\) and also on the parametric families used for modeling the lifetimes.
Similarly, alternative optimization methods such as the algorithm LRS discussed in Sect. 4, though being theoretically basic, can be valuable in certain situations, particularly when initial values of the Newton–Raphson methods are too far from the optimum. Owing to the relatively robust search strategy, the algorithm LRS can be used as a preparation for more sophisticated optimization methods such as the algorithm MNS.
All three considered algorithms require initial values \(\varvec{\theta }_j^0, j=1, \ldots , m\), for which we suggest using the conventional maximum likelihood estimate in the absence of truncation. Though it has not been further examined in the simulation study, this choice has generally lead to surprisingly satisfactory results. Alternative initial values might yield better numerical performance under certain circumstances.
We have sketched how the asymptotic behaviour of the estimates \(\varvec{\widehat{a}}\) and \(\varvec{\widehat{\varvec{\theta }}}\) can be studied for given consistent estimator \(\widehat{G}^{(n)}\). It remains to prove that the proposed nonparametric estimator \(\widehat{G}^{(n)}\) is in fact consistent.
Availability of data and material (data transparency)
The data were collected from a public website.
References
Chiou, S. H., Austin, M. D., Qian, J., & Betensky, R. A. (2019). Transformation model estimation of survival under dependent truncation and independent censoring. Statistical Methods in Medical Research, 28(12), 3785–3798. https://doi.org/10.1177/0962280218817573.
Chiou, S. H., Qian, J., Mormino, E., & Betensky, R. A. (2018). Permutation tests for general dependent truncation. Computational Statistics and Data Analysis, 128, 308–324. https://doi.org/10.1016/j.csda.2018.07.012.
de Uña-Álvarez, J. (2020). Nonparametric estimation of the cumulative incidences of competing risks under double truncation. Biometrical Journal, 62(3), 852–867. https://doi.org/10.1002/bimj.201800323.
Dörre, A. (2020). Bayesian estimation of a lifetime distribution under double truncation caused by time-restricted data collection. Statistical Papers, 61(3), 941–965. https://doi.org/10.1007/s00362-017-0968-7.
Dörre, A., & Emura, T. (2019). Analysis of doubly truncated data: An introduction. Berlin: Springer.
Efron, B., & Petrosian, V. (1999). Nonparametric methods for doubly truncated data. Journal of the American Statistical Association, 94(447), 824–834.
Emura, T., Hu, Y., & Konno, Y. (2017). Asymptotic inference for maximum likelihood estimators under the special exponential family with double-truncation. Statistical Papers, 58, 877–909. https://doi.org/10.1007/s00362-015-0730-y.
Emura, T., & Konno, Y. (2012). Multivariate normal distribution approaches for dependently truncated data. Statistical Papers, 53(1), 133–149.
Emura, T., Konno, Y., & Michimae, H. (2015). Statistical inference based on the nonparametric maximum likelihood estimator under double-truncation. Lifetime Data Analysis, 21(3), 397–418.
Lagakos, S. W., Barraj, L. M., & de Gruttola, V. (1988). Nonparametric analysis of truncated survival data with application to AIDS. Biometrika, 75(3), 515–523.
Lawless, J. F., & Kalbfleisch, J. D. (1992). Some issues in the collection and analysis of field reliability data. In J. P. Klein & P. K. Goel (Eds.), Survival analysis: State of the Art. Nato Science (series E: applied sciences) (Vol. 211, pp. 141–152). Dordrecht: Springer.
Martin, E. C., & Betensky, R. A. (2005). Testing quasi-independence of failure and truncation times via conditional Kendall’s tau. Journal of the American Statistical Association, 100(470), 484–492.
Moreira, C., & de Uña-Álvarez, J. (2010). Bootstrapping the NPMLE for doubly truncated data. Journal of Nonparametric Statistics, 22, 567–583.
Moreira, C., de Uña-Álvarez, J., & Braekers, R. (2021). Nonparametric estimation of a distribution function from doubly truncated data under dependence. Computational Statistics,. https://doi.org/10.1007/s00180-021-01085-4.
Moreira, C., de Uña-Álvarez, J., & van Keilegom, I. (2014). Goodness-of-fit tests for a semiparametric model under random double truncation. Computational Statistics, 29, 1365–1379. https://doi.org/10.1007/s00180-014-0496-z.
Reiss, R.-D. (1993). A course on point processes. New York: Springer.
Scheike, T. H., & Keiding, N. (2006). Design and analysis of time-to-pregnancy. Statistical Methods in Medical Research, 15, 127–140.
Shen, P. (2010a). Nonparametric analysis of doubly truncated data. Annals of the Institute of Statistical Mathematics, 62(5), 835–853.
Shen, P. (2010b). Semiparametric analysis of doubly truncated data. Communications in Statistics Theory and Methods, 39(17), 3178–3190. https://doi.org/10.1080/03610920903219272.
Snyder, D. L., & Miller, M. I. (1991). Random point processes in time and space (2nd ed.). Berlin: Springer.
Tsai, W. Y. (1990). Testing the assumption of independence of truncation time and failure time. Biometrika, 77(1), 169–177.
van der Vaart, A. W. (1998). Asymptotic statistics. Cambridge University Press. https://doi.org/10.1017/CBO9780511802256.
Wang, M.-C. (1989). A semiparametric model for randomly truncated data. Journal of the American Statistical Association, 84(407), 742–748.
Xiao, J., & Hudgens, M. G. (2019). On nonparametric maximum likelihood estimation with double truncation. Biometrika, 106(4), 989–996.
Ye, Z., & Tang, L. (2016). Augmenting the unreturned for field data with information on returned failures only. Technometrics, 58(4), 513–523.
Zhang, X. (2015). Nonparametric inference for an inverse-probability-weighted estimator with doubly truncated data. Communications in Statistics Simulation and Computation, 44(2), 489–504.
Zhigljavsky, A. A. (1991). Theory of global random search. Berlin: Springer Science + Business.
Acknowledgements
The author is grateful to Takeshi Emura for the invitation to contribute to the special issue and for numerous inspiring discussions on the addressed topic. For their many constructive and effective comments, the author wishes to express his gratitude to the anonymous reviewers. He also thanks Rafael Weißbach and Jacobo de Uña-Álvarez for many stimulating conversations.
Funding
Open Access funding enabled and organized by Projekt DEAL. No particular funding has been received for this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There are no competing interests.
Code availability (software application or custom code)
Implementation has been performed as custom code and can be made publicly available.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Appendix
Appendix

As in the algorithm MNS, we use the maximum likelihood estimates of \(\varvec{\theta }_j\) in the model without truncation for the initial values \(\varvec{\theta }_j^0\). In Step 2, the parameters \(q_G, q_{{\varvec{a}}}, q_{\varvec{\theta }} \in (0, 1)\) define the maximum relative step size for each parameter and iteration. An efficient choice of these values ensures a sufficiently large probability of finding parameter values with increased log-likelihood value, and a sufficiently large average step size to enable a feasible runtime by avoiding unnecessarily many iterations. An optimal choice generally depends on the setup, in particular the sample size and the considered lifetime distributions. In the studied simulation setup, we have found that choosing \(q_G = n^{-1}, q_{{\varvec{a}}} = 0.05\) and \(q_{\varvec{\theta }} = 0.025\) seems to be a suitable practical choice for small to moderate sample sizes.

The damping factor \(\omega \) is incorporated as in the algorithm MNS. For sample sizes \(n\approx 1500\), a damping factor of \(\omega = 0.25\) seemed suitable for both avoiding invalid parameter values and still achieving satisfactory runtime of the algorithm. Note that in Step 2, the current parameter \({\widetilde{\varvec{\gamma }}}\) is adjusted in each univariate Newton–Raphson step. As in the algorithm MNS, we use the maximum likelihood estimates of \(\varvec{\theta }_j\) in the model without truncation for the initial values \(\varvec{\theta }_j^0\).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dörre, A. Semiparametric likelihood inference for heterogeneous survival data under double truncation based on a Poisson birth process. Jpn J Stat Data Sci 4, 1203–1226 (2021). https://doi.org/10.1007/s42081-021-00128-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42081-021-00128-w