Abstract
Electroencephalography (EEG) is a non-invasive way of recording brain activities, making it useful for diagnosing various neurological disorders. However, artifact signals associated with eye blinks or the heart spread across the scalp, contaminating EEG recordings and making EEG data analysis difficult. To solve this problem, we implement a common methodology to suppress both cardiac and ocular artifact signal, by correlating the measured contaminated EEG signals with the clean reference electro-oculography (EOG) and electrocardiography (EKG) data and subtracting the scaled EOG and EKG from the contaminated EEG recording. In the proposed methodology, the clean EOG and EKG signals are extracted by subjecting the raw reference time-series data to ensemble empirical mode decomposition to obtain the intrinsic mode functions. Then, an unsupervised technique is used to capture the artifact components. We compare the distortion introduced into the brain signal after artifact suppression using the proposed method with those obtained using conventional regression alone and with a wavelet-based approach. The results show that the proposed method outperforms the other techniques, with an additional advantage of being a common methodology for the suppression of two types of artifact.
Similar content being viewed by others
1 Introduction
Electroencephalography (EEG) is an electrophysiological monitoring method for recording the electrical activity of the brain, giving insight into the brain and its abnormalities. EEG signals are recorded from the scalp either with electrodes in the standard 10–20 arrangement or an extended 10–20 system (64 electrodes), following a standard devised by the International Federation of Societies of EEG [1]. EEG signals are often contaminated by potentials of non-cerebral origin, such as the heart and eyeblinks; these signals pose a serious problem in analyzing the recorded EEG activity [2, 3]. A number of techniques, such as principal component analysis (PCA) [4], independent component analysis (ICA) [5, 6], wavelet transform [7–10], and regression [11] have been suggested for eliminating the relatively common ocular artifacts and somewhat rare artifacts due to activity [12]. Artifact suppression techniques based on statistical methods such as ICA have been used for artifact rejection [13, 14]. The main and inevitable problem of ICA methodology is that the independent components attributed to the artifacts may also contain neural activity of interest. In this sense, the omission of particular components corresponding to the artifacts, followed by a signal reconstruction from the remaining components, may lead to distortions of the underlying cerebral activity [15]. Wavelet-based approaches are a relatively easier technique for artifact suppression and can also be used for single-channel EEG data. However, it is well known that wavelet-based methods require a set of pre-defined basis functions to decompose the signal and that the selection of a particular basis function is governed by morphological features of the signal to be decomposed. Hence, a common basis function may not be optimal for the suppression of artifacts associated with different sources (eyeblinks and the heart) using a wavelet-based approach.
Recently, some researchers [16–18] have used data-driven techniques such as empirical mode decomposition (EMD) and ensemble empirical mode decomposition (EEMD) for ocular artifact suppression; however, these works did not address the development of a single or common methodology for the suppression of artifacts associated with both ocular and cardiac activities.
Regression-based methodology is one of the simplest techniques for removing ocular artifacts, where the measured EOG signal is projected onto the EEG channels. However, this method might produce unexpected distortion in the clean EEG data due to bidirectional contamination [2]. To overcome the bidirectional contamination problem inherent in the regression approach, we propose performing EEMD with an unsupervised technique on the raw reference data to obtain the clean reference data. Here, we propose a common methodology for the suppression of two kinds of biological artifact (EOG and EKG) from the contaminated EEG by using a regression technique between the measured contaminated EEG and the clean EKG and EOG reference data.
The rest of this paper is organized as follows. Section 2 presents a brief introduction to regression and EEMD. Section 3 introduces the experimental protocol as well as an overview of existing approaches for artifact suppression and their comparative evaluation, followed by artifact suppression using the proposed technique. Section 4 presents the results and the metrics used to evaluate and compare the performance of the proposed methodology with other techniques. Finally, a discussion and the conclusion are presented in Sect. 5.
2 Regression and EEMD
In the conventional regression technique, ocular and cardiac artifacts are suppressed by projecting the measured raw reference signals related to EOG and EKG onto the EEG data. However, the regression technique does not take into account bidirectional contamination, i.e., the EOG recording is also corrupted by cerebral activity, and thus this cerebral activity will also be eliminated from the EEG recordings after ocular artifact suppression using the conventional regression-based technique. Low-pass filtering has been suggested for reducing the suppression of high-frequency cerebral components from EEG data [19]; however, the criterion for the selection of the cut-off frequency was not discussed.
The EMD technique adaptively decomposes non-stationary signals into a set of intrinsic oscillatory modes termed as intrinsic mode functions (IMFs) [20]. These IMFs represent signal components with progressively decreasing frequency content. The EMD algorithm applied to a given data x(t)for deriving the IMFs is defined in the following way.
The EMD technique adaptively decomposes non-stationary signals into a set of intrinsic oscillatory modes, called intrinsic mode functions (IMFs) [20]. These IMFs represent signal components with progressively decreasing frequency content. The EMD algorithm applied to a given data x(t) for deriving IMFs is defined as follows:
-
1.
All the extrema in the data are identified and connected by a cubic spline to form upper and lower envelopes.
-
2.
The mean of the upper and lower envelopes m 1(t) is obtained and the mean from the original series is subtracted to get h 1(t) as:
$$h_{1} \left( t \right) = x\left( t \right) - m_{1} \left( t \right)$$(1) -
3.
If the difference h 1(t) does not satisfy the IMF conditions, steps (1) and (2) are repeated until the envelopes are symmetric with respect to zero under standard stopping criteria, described elsewhere [20, 21].
-
4.
When the desired stopping criterion is achieved, the first IMF, c 1(t), is obtained and the residue is calculated as:
$$r_{1} \left( t \right) = x\left( t \right) - c_{1} \left( t \right)$$(2) -
5.
Now, residue r 1 (t) is treated as the signal to be further decomposed and the above steps are repeated to obtain r 2(t), r 3(t),…,r m(t).
The original signal can be represented as,
where c i(t) is the ith order IMF and r m(t) is the residue or the last IMF.
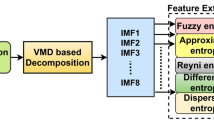
Although the adaptive EMD method is useful for non-linear and non-stationary signals, it has the disadvantage of a mode mixing effect, which arises due to the inter-mittency present in the signal. To overcome this, an EEMD method was suggested to prevent mode mixing in IMFs [22]. To avoid this mode mixing, a finite amplitude of identically distributed white noise is added to the input signal before the EMD is performed, and the ensemble average of IMFs is taken over a number of trials to avoid the mode mixing effect. The noise added in each trial tends to cancel when the ensemble average is taken as there is no correlation between the noise introduced in different trials. In our case, the noise amplitude added to EEMD is 0.3 times the standard deviation and the number of ensemble averages is fixed at 100.
3 Method
3.1 Data Acquisition
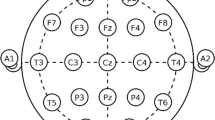
A total of four male subjects (28–45 years old) took part in the EEG experiments. Written informed consent for participation was obtained from all the subjects. A 64-channel EEG system (Compumedics Neuroscan Synamps 64 Channel EEG System, Australia) was used for acquiring EEG signals from the brain. The system is equipped with a data acquisition card with 24-bit resolution. EEG recordings were carried out with a sampling rate of 1 kHz and the allowable bandwidth set from 0 to 200 Hz. The EEG data were recorded for 2 min per subject. Separate electrodes (EOG and EKG) were also used to acquire eye blinks and heart activity simultaneously with the EEG. EOG artifacts were observed in most of the EEG recordings during the pattern reversal checkerboard EEG experiments, but EKG artifacts were observed for only one male subject. For data analysis, five EEG data segments of 2.5 s contaminated by eyeblinks were taken from every subject and five EEG data segments of 1 s contaminated by cardiac activity were selected from a male subject.
3.2 Artifact Removal Evaluation
The aim of the artifact suppression algorithm is to selectively suppress artifacts without much affecting the signal from the brain. To compare the performance of the artifact suppression technique, we calculated the change in power spectral density (ΔPSD) before and after performing artifact suppression [23]. PSD is a measure of signal power in the selected frequency domain. In our case, we calculated the change in signal power (ΔPSD) over a contaminated EEG data segment before and after artifact suppression using the Welch’s method across the frequency bands corresponding to different brain rhythms, i.e., delta (0.5–4 Hz), theta (4–8 Hz), alpha (8–12 Hz), and beta (13–30 Hz); the respective changes in PSD are denoted as ΔPSDδ, ΔPSDθ, ΔPSDα, and ΔPSDβ. These metrics were used for the quantitative evaluation of the distortion introduced in the EEG signals after artifact suppression. Lower values of ΔPSD correspond to lower distortion introduced in the underlying cerebral activity by the algorithm used for artifact suppression.
3.3 Data Analysis
EOG interference was dominant in the frontal lobe electrodes FP2, FP1, and FZ, whereas EKG interference was dominant in the central lobe electrodes CP2, CP1, C1, and C2. Samples belonging to EOG and EKG recordings and the corresponding contaminated part of EEG from the frontal FP2 electrode and the central CP2 electrode were taken off from the raw EEG recording for analysis, as shown in Figs. 1 and 2. EEG data sets were converted from the continuous format (.cnt) to ASCII format (.dat) to make use of Python for further analysis [24–26]. The applied methodology is summarized in the following steps:
-
1.
EEMD was performed on the raw EOG reference electrode data, as shown in Figs. 3 and 4, to get the IMFs. Similarly, the raw EKG data were also subjected to EEMD to get the IMFs, as shown in Figs. 5 and 6. Then, an unsupervised technique called PCA was used to capture the principal components for extracting eyeblinks and cardiac activity.
Fig. 4 -
2.
The number of principal components retained for the reconstruction of the clean EOG and EKG reference data is based on the threshold value driven from the scree plot [27]. Figures 7 and 8 show the clean EOG and EKG reference data, respectively, after applying the above approach on the raw reference data.
Fig. 7 -
3.
After extracting the clean reference data, correlations were calculated between contaminated EEG and the clean reference data (EOG and EKG), and then the scaled EOG and EKG data were subtracted from the corresponding contaminated EEG recording in order to suppress ocular and cardiac artifacts.
As with eyeblinks, heart and brain activities have physiologically distinct sources. The measured EEG is thus a superposition of the true EEG signal and a scaled part of the clean EOG or EKG signal. The contaminated recorded EEG data can be represented as:
where EEG1mes = measured contaminated EEG at frontal lobe(FP2), EEG2mes = measured contaminated EEG at central lobe(CP2), EEG1true and EEG2true = EEG due to cortical (brain) activity alone at measurement electrode, kEOG·EOGcleaned = propagated EOG artifact from eye to recording site, kEKG·EKGcleaned = propagated EKG artifact from heart to recording site.
EEG1true and EEG2true signals were estimated from EEG1mes and EEG2mes by efficiently removing the kEOG ·EOGclean and kEKG·EKGclean while retaining the EEG1true and EEG2true signal integrity. It is necessary to determine the coefficients kEOG and kEKG for extracting the EEG1true and EEG2true signals. To determine kEOG and kEKG, the covariance between EEG1mes and EOGclean and between EEG2mes and EKGclean, and the variance for EOGclean and EKGclean were calculated.
The scale factors kEOG and kEKGwere respectively estimated as:
Finally, EEG1true and EEG2true signals were estimated using the coefficients kEOG and kEKG in Eqs. (3) and (4).
4 Results
Figures 9 and 10 show the clean and contaminated EEG data obtained using the proposed technique. From their visual appearance, it is evident that the proposed methodology worked well for the effective suppression of EOG and EKG artifacts. In order to compare the performance of the proposed method with a conventional wavelet-based technique, we extracted the signal corresponding to eyeblinks and cardiac activity from the contaminated EEG data segment using Biorthogonal 3.3 [28] and Symlet (sym8) [29] wavelets, respectively. In our case, since the measurement bandwidth allowed is 0–200 Hz, decomposition up to the fourth level was selected as the range of frequency for ocular artifacts is 0-10 Hz [30].For cardiac artifact suppression, decomposition up to the second level was performed. To compare the results obtained using the wavelet-based approach for the suppression of artifacts, we reconstructed clean EEG data from the contaminated EEG data by setting the noisy coefficients of the wavelet-decomposed data to zero. Tables 1 and 2 indicate that significantly lower distortion in the brain signal results when the proposed technique is used for artifact suppression compared to those obtained with the conventional regression alone and the wavelet-based approach.
5 Conclusion
We proposed a common methodology for the suppression of artifacts associated with heart activity and eyeblinks from measured EEG data by calculating and subtracting the relative changes in the EEG data with respect to the clean reference EKG and EOG data. The results show that the proposed methodology is able to eliminate EOG and EKG artifacts from the EEG data while causing little distortion in the underlying cerebral activity. Compared to other artifact suppression techniques, the proposed method has several advantages. It can be used for the suppression of artifacts associated with different sources (eyeblinks and the heart) using single-channel contaminated EEG data along with measured reference electrode EOG/EKG data, whereas PCA and ICA methods require multi-channel EEG data. The proposed method overcomes the problem of bidirectional contamination between EEG and an EOG channel that is difficult to properly resolve using the conventional regression-based approach. The significance of the proposed method lies in the fact that it is simple and easy to implement, unlike wavelet- and ICA-based artifact suppression approaches, which require careful selection of optimal mother wavelet function and noisy components, respectively.
Change history
10 February 2018
The article “Common Methodology for Cardiac and Ocular Artifact Suppression from EEG Recordings by Combining Ensemble Empirical Mode Decomposition with Regression Approach”, written by Rajesh Patel, K. Gireesan, S. Sengottuvel, M. P. Janawadkar, T. S. Radhakrishnan was originally published Online First without open access. After publication in volume [37], issue [2], page [201-208] the author decided to opt for Open Choice and to make the article an open access publication. Therefore, the copyright of the article has been changed to © The Author(s) [2018] and the article is forthwith distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, duplication, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made
10 February 2018
The article ���Common Methodology for Cardiac and Ocular Artifact Suppression from EEG Recordings by Combining Ensemble Empirical Mode Decomposition with Regression Approach���, written by Rajesh Patel, K. Gireesan, S. Sengottuvel, M. P. Janawadkar, T. S. Radhakrishnan was originally published Online First without open access. After publication in volume [37], issue [2], page [201-208] the author decided to opt for Open Choice and to make the article an open access publication. Therefore, the copyright of the article has been changed to �� The Author(s) [2018] and the article is forthwith distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, duplication, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made
References
Jasper, H. H. (1958). Report of the committee on methods of clinical examination in electroencephalography. Electroencephalography and Clinical Neurophysiology, 10, 370–375.
Urigen, J. A., & Garcia-Zapirain, B. (2015). EEG artifact removal: State-of-the-art and guidelines. Journal of Neural Engineering, 12(3), 031001. http://stacks.iop.org/1741-2552/12/i=3/a=031001.
Vigon, L., Saatchi, M. R., Mayhew, J. E. W., & Fernandes, R. (2000). Quantitative evaluation of techniques for ocular artefact filtering of EEG waveforms. IEEE Proceedings—Science, Measurement and Technology, 147, 219–228.
Jung, T. P., Makeig, S., Humphries, C., Lee, T.-W., McKeown, M. J., Iragui, V., et al. (2000). Removing electroencephalographic artifacts by blind source separation. Psychophysiology, 37(2), 163–178.
Comon, P. (1994). Independent component analysis, a new concept? Signal Processing, 36(3), 287–314.
Romero, S., Maanas, M., & Barbanoj, M. (2009). Ocular reduction in EEG signals based on adaptive filtering, regression and blind source separation. Annals of Biomedical Engineering, 37(1), 176–191.
Zikov, T., Bibian, S., Dumont, G. A., Huzmezan, M.,& Ries, C. R. (2002). A wavelet based de-noising technique for ocular artifact correction of the electroencephalogram. In 24th Annual conference and the annual fall meeting of the Biomedical Engineering Society EMBS/BMES conference. Proceedings of the second joint engineering in medicine and biology (Vol. 1, pp. 98–105).
Krishnaveni, V., Jayaraman, S., Aravind, S., Hariharasudhan, V., & Ramadoss, K. (2006). Automatic identification and removal of ocular artifacts from EEG using wavelet transform. Measurement Science Review, 6(4), 45–57.
Hsu, W. Y., Lin, C. H., Hsu, H. J., Chen, P. H., & Chen, I.-R. (2012). Wavelet-based envelope features with automatic EOG artifact removal: Application to single-trial EEG data. Expert Systems with Applications, 39(3), 2743–2749.
Al-Qazzaz, N., Ali, S., Ahmad, S., Islam, M., & Ariff, M. (2014). Selection of mother wavelets thresholding methods in denoising multi-channel EEG signals during working memory task. In 2014 IEEE conference on biomedical engineering and sciences (IECBES) (pp. 214–219).
He, P., Wilson, G., Russell, C., & Gerschutz, M. (2007). Removal of ocular artifacts from the EEG: A comparison between time-domain regression method and adaptive filtering method using simulated data. Medical & Biological Engineering & Computing, 45(5), 495–503.
Devuyst,S., Dutoit, T., Stenuit, P., Kerkhofs, M., & Stanus, E. (2008). Removal of ECG artifacts from EEG using a modified independent component analysis approach. In Engineering in Medicine and Biology Society, 2008. EMBS 2008. 30th Annual international conference of the IEEE (pp. 5204–5207).
Hamaneh, M., Chitravas, N., Kaiboriboon, K., Lhatoo, S., & Loparo, K. (2014). Automated removal of EKG artifact from EEG data using independent component analysis and continuous wavelet transformation. IEEE Transactions on Biomedical Engineering, 61(6), 1634–1641.
Mahajan, R., & Morshed, B. (2015). Unsupervised eye blink artifact denoising of EEG data with modified multiscale sample entropy, kurtosis, and wavelet-ICA. IEEE Journal of Biomedical and Health Informatics, 19(1), 158–165.
Castellanos, N. P., & Makarov, V. A. (2006). Recovering EEG brain signals: Artifact suppression with wavelet enhanced independent component analysis. Journal of Neuroscience Methods, 158(2), 300–312.
Shahbakhti,M., Khalili, V., & Kamaee, G. (2012). Removal of blink from EEG by empirical mode decomposition (EMD). In Biomedical engineering international conference (BMEiCON) (pp. 1–5).
Zeng, H., Song, A., Yan, R., & Qin, H. (2013). EOG artifact correction from EEG recording using stationary subspace analysis and empirical mode decomposition. Sensors, 13(11), 14839–14859.
Patel, R., Sengottuvel, S., Janawadkar, M., Gireesan, K., Radhakrishnan, T., & Mariyappa, N. (2015). Ocular artifact suppression from EEG using ensemble empirical mode decomposition with principal component analysis. Computers and Electrical Engineering, 54, 78–86.
Gasser, T., Ziegler, P., & Gattaz, W. (1992). The deleterious effect of ocular artefacts on the quantitative EEG, and a remedy. European Archives of Psychiatry and Clinical Neuroscience, 241(6), 352–356.
Huang, N. E., Shen, Z., Long, S. R., Wu, M. C., Shih, H. H., Zheng, Q., Yen, N.-C., Tung, C. C., & Liu, H. H. (1998). The empirical mode decomposition and the Hilbert spectrum for nonlinear and non-stationary time series analysis. Proceedings of the Royal Society of London A: Mathematical, Physical and Engineering Sciences, 454, 903–995. http://rspa.royalsocietypublishing.org/content/454/1971/903.
Rilling, G., Flandrin, P., & Goncalves, P. (2003). On empirical mode decomposition and its algorithms. In IEEE-EURASIP workshop on nonlinear signal and image processing NSIP03.
Wu, Z., & Huang, N. E. (2009). Ensemble empirical mode decomposition: A noise-assisted data analysis method. Advances in Adaptive Data Analysis, 01(01), 1–41.
Klados, M. A., Papadelis, C., Braun, C., & Bamidis, P. D. (2011). REG-ICA: A hybrid methodology combining blind source separation and regression techniques for the rejection of ocular artifacts. Biomedical Signal Processing and Control, 6(3), 291–300.
Perez, F., & Granger, B. E. (2007). IPython: A system for interactive scientific computing. Computing in Science Engineering, 9(3), 21–29.
Sederberg, P. B., & Weidemann, C. T. (2007). EMD code along with the Python time series analysis. http://github.com/compmem/ptsa/blob/master/ptsa/emd.py.
Hunter, J. D. (2007). Matplotlib: A 2D graphics environment. Computing in Science Engineering, 9(3), 90–95.
Zhou, L., & Thomas, R. D. (2015). Principal component analysis of the memory load effect in a change detection task. Vision Research, 110(Part A), 1–6.
Naga, R., Chandralingam, S., Anjaneyulu, T., & Satyanarayana, K. (2012). Denoising EOG signal using stationary wavelet transform. Measurement Science Review, 12(2), 46–51.
Chouakri, S., Bereksi-Reguig, F., Ahmaidi, S., & Fokapu, O. (2005). Wavelet denoising of the electro-cardiogram signal based on the corrupted noise estimation. Computers in Cardiology, 2005, 1021–1024.
Zhou, W., & Gotman, J. (2009). Automatic removal of eye movement artifacts from the EEG using ICA and the dipole model. Progress in Natural Science, 19(9), 1165–1170.
Acknowledgements
The authors would like to thank Dr. G. Amarendra, Director MSG, IGCAR, for his valuable support throughout this research work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This study was carried out on volunteers (author and co-authors), with the informed consent obtained from them.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Patel, R., Gireesan, K., Sengottuvel, S. et al. Common Methodology for Cardiac and Ocular Artifact Suppression from EEG Recordings by Combining Ensemble Empirical Mode Decomposition with Regression Approach. J. Med. Biol. Eng. 37, 201–208 (2017). https://doi.org/10.1007/s40846-016-0208-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40846-016-0208-y