Abstract
Purpose of Review
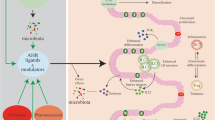
Intestinal stem cells, the most rapidly proliferating adult stem cells, are exquisitely sensitive to extrinsic dietary factors. Uncontrolled regulation of intestinal stem cells is closely linked to colon tumorigenesis. This review focuses on how dietary- and microbial-derived cues regulate intestinal stem cell functionality and colon tumorigenesis in mouse models by targeting the aryl hydrocarbon receptor (AhR).
Recent Findings
AhR, a ligand-activated transcription factor, can integrate environmental, dietary, and microbial cues to modulate intestinal stem cell proliferation, differentiation, and their microenvironment, affecting colon cancer risk. Modulation of AhR activity is associated with many chronic diseases, including inflammatory bowel diseases where AhR expression is protective.
Summary
AhR signaling controls the maintenance and differentiation of intestinal stem cells, influences local niche factors, and plays a protective role in colon tumorigenesis. Mounting evidence suggests that extrinsic nutritional/dietary cues which modulate AhR signaling may be a promising approach to colon cancer chemoprevention.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Barker N, Clevers H. Tracking down the stem cells of the intestine: strategies to identify adult stem cells. Gastroenterology. 2007;133(6):1755–60. https://doi.org/10.1053/j.gastro.2007.10.029.
Carmon KS, Lin Q, Gong X, Thomas A, Liu Q. LGR5 interacts and cointernalizes with Wnt receptors to modulate Wnt/beta-catenin signaling. Mol Cell Biol. 2012;32(11):2054–64. https://doi.org/10.1128/MCB.00272-12.
Barker N, Clevers H. Lineage tracing in the intestinal epithelium. Current protocols in stem cell biology. 2010;Chapter 5:Unit5A.4. https://doi.org/10.1002/9780470151808.sc05a04s13.
Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457(7229):608–11. https://doi.org/10.1038/nature07602.
Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–5. https://doi.org/10.1038/nature07935.
Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143(1):134–44. https://doi.org/10.1016/j.cell.2010.09.016.
Qiu W, Wang X, Leibowitz B, Liu H, Barker N, Okada H, et al. Chemoprevention by nonsteroidal anti-inflammatory drugs eliminates oncogenic intestinal stem cells via SMAC-dependent apoptosis. Proc Natl Acad Sci U S A. 2010;107(46):20027–32. https://doi.org/10.1073/pnas.1010430107.
Leung C, Tan SH, Barker N. Recent advances in Lgr5(+) stem cell research. Trends Cell Biol. 2018;28(5):380–91. https://doi.org/10.1016/j.tcb.2018.01.010.
Kim C-K, Yang VW, Bialkowska AB. The role of intestinal stem cells in epithelial regeneration following radiation-induced gut injury. Curr Stem Cell Rep. 2017;3(4):320–32. https://doi.org/10.1007/s40778-017-0103-7.
Vermeulen L, Snippert HJ. Stem cell dynamics in homeostasis and cancer of the intestine. Nat Rev Cancer. 2014;14(7):468–80. https://doi.org/10.1038/nrc3744.
McCarthy N, Kraiczy J, Shivdasani RA. Cellular and molecular architecture of the intestinal stem cell niche. Nat Cell Biol. 2020;22(9):1033–41. https://doi.org/10.1038/s41556-020-0567-z.
García-Prat L, Sousa-Victor P, Muñoz-Cánoves P. Proteostatic and metabolic control of stemness. Cell Stem Cell. 2017;20(5):593–608. https://doi.org/10.1016/j.stem.2017.04.011.
Ren R, Ocampo A, Liu GH, Izpisua Belmonte JC. Regulation of stem cell aging by metabolism and epigenetics. Cell Metab. 2017;26(3):460–74. https://doi.org/10.1016/j.cmet.2017.07.019.
Schell JC, Wisidagama DR, Bensard C, Zhao H, Wei P, Tanner J, et al. Control of intestinal stem cell function and proliferation by mitochondrial pyruvate metabolism. Nat Cell Biol. 2017;19(9):1027–36. https://doi.org/10.1038/ncb3593.
Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23(10):1124–34. https://doi.org/10.1038/nm.4409.
Alonso S, Yilmaz ÖH. Nutritional regulation of intestinal stem cells. Annu Rev Nutr. 2018;38(1):273–301. https://doi.org/10.1146/annurev-nutr-082117-051644.
Lee Chong T, Ahearn EL, Cimmino L. Reprogramming the epigenome with vitamin C. Front Cell Dev bIol. 2019;7(128). https://doi.org/10.3389/fcell.2019.00128.
Xing PY, Pettersson S, Kundu P. Microbial metabolites and intestinal stem cells tune intestinal homeostasis. PROTEOMICS. 2020;20(5–6):1800419. https://doi.org/10.1002/pmic.201800419.
Kim E, Davidson LA, Zoh RS, Hensel ME, Salinas ML, Patil BS, et al. Rapidly cycling Lgr5(+) stem cells are exquisitely sensitive to extrinsic dietary factors that modulate colon cancer risk. Cell Death Dis. 2016;7(11):e2460. https://doi.org/10.1038/cddis.2016.269.
Kim E, Wright GA, Zoh RS, Patil BS, Jayaprakasha GK, Callaway ES, et al. Establishment of a multicomponent dietary bioactive human equivalent dose to delete damaged Lgr5+ stem cells using a mouse colon tumor initiation model. European journal of cancer prevention : the official journal of the European Cancer Prevention Organisation (ECP). 2019;28(5):383–9. https://doi.org/10.1097/cej.0000000000000465.
Fan YY, Davidson LA, Callaway ES, Wright GA, Safe S, Chapkin RS. A bioassay to measure energy metabolism in mouse colonic crypts, organoids, and sorted stem cells. Am J Physiol Gastrointest Liver Physiol. 2015;309(1):G1–9. https://doi.org/10.1152/ajpgi.00052.2015.
Han H, Davidson LA, Fan Y-Y, Goldsby JS, Yoon G, Jin U-H, et al. Loss of aryl hydrocarbon receptor potentiates FoxM1 signaling to enhance self-renewal of colonic stem and progenitor cells. EMBO J. 2020:e104319. https://doi.org/10.15252/embj.2019104319Demonstrates that AhR signaling directly controls the expression of FoxM1 to regulate the functionality of colonic stem/progenitor cells, thus affecting colon tumorigenesis.
Bradfield CA, Poland A. A competitive binding assay for 2,3,7,8-tetrachlorodibenzo-p-dioxin and related ligands of the Ah receptor. Mol Pharmacol. 1988;34(5):682–8.
Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, Trump S, et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478(7368):197–203. https://doi.org/10.1038/nature10491.
Hubbard TD, Murray IA, Bisson WH, Lahoti TS, Gowda K, Amin SG, et al. Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci Rep. 2015;5:12689. https://doi.org/10.1038/srep12689.
Jin UH, Karki K, Cheng Y, Michelhaugh SK, Mittal S, Safe S. The aryl hydrocarbon receptor is a tumor suppressor-like gene in glioblastoma. J Biol Chem. 2019;294(29):11342–53. https://doi.org/10.1074/jbc.RA119.008882.
Soshilov A, Denison MS. Role of the Per/Arnt/Sim domains in ligand-dependent transformation of the aryl hydrocarbon receptor. J Biol Chem. 2008;283(47):32995–3005. https://doi.org/10.1074/jbc.M802414200.
Denison MS, Soshilov AA, He G, DeGroot DE, Zhao B. Exactly the same but different: promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol Sci. 2011;124(1):1–22. https://doi.org/10.1093/toxsci/kfr218.
Poland A, Glover E. Stereospecific, high affinity binding of 2,3,7,8-tetrachlorodibenzo-p-dioxin by hepatic cytosol. Evidence that the binding species is receptor for induction of aryl hydrocarbon hydroxylase. J Biol Chem. 1976;251(16):4936–46.
Lucier GW, McDaniel OS, Hook GE, Fowler BA, Sonawane BR, Faeder E. TCDD-induced changes in rat liver microsomal enzymes. Environ Health Perspect. 1973;5:199–209. https://doi.org/10.1289/ehp.7305199.
Li S, Pei X, Zhang W, Xie HQ, Zhao B. Functional analysis of the dioxin response elements (DREs) of the murine CYP1A1 gene promoter: beyond the core DRE sequence. Int J Mol Sci. 2014;15(4):6475–87. https://doi.org/10.3390/ijms15046475.
Davarinos NA, Pollenz RS. Aryl hydrocarbon receptor imported into the nucleus following ligand binding is rapidly degraded via the cytosplasmic proteasome following nuclear export. J Biol Chem. 1999;274(40):28708–15. https://doi.org/10.1074/jbc.274.40.28708.
Huang G, Elferink CJ. A novel nonconsensus xenobiotic response element capable of mediating aryl hydrocarbon receptor-dependent gene expression. Mol Pharmacol. 2012;81(3):338–47. https://doi.org/10.1124/mol.111.075952.
Wilson SR, Joshi AD, Elferink CJ. The tumor suppressor Kruppel-like factor 6 is a novel aryl hydrocarbon receptor DNA binding partner. J Pharmacol Exp Ther. 2013;345(3):419–29. https://doi.org/10.1124/jpet.113.203786.
Ohtake F, Baba A, Takada I, Okada M, Iwasaki K, Miki H, et al. Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature. 2007;446(7135):562–6. https://doi.org/10.1038/nature05683.
Kawajiri K, Kobayashi Y, Ohtake F, Ikuta T, Matsushima Y, Mimura J, et al. Aryl hydrocarbon receptor suppresses intestinal carcinogenesis in ApcMin/+ mice with natural ligands. Proc Natl Acad Sci U S A. 2009;106(32):13481–6. https://doi.org/10.1073/pnas.0902132106.
Shiizaki K, Kido K, Mizuta Y. Insight into the relationship between aryl-hydrocarbon receptor and β-catenin in human colon cancer cells. PLOS ONE. 2019;14(11):e0224613. https://doi.org/10.1371/journal.pone.0224613Provides follow-up evidence that AhR does not act as a E3 ligase to mediate β-catenin degradation in human colon cancer cell lines.
Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14(8):e1002533. https://doi.org/10.1371/journal.pbio.1002533.
Jin UH, Lee SO, Sridharan G, Lee K, Davidson LA, Jayaraman A, et al. Microbiome-derived tryptophan metabolites and their aryl hydrocarbon receptor-dependent agonist and antagonist activities. Mol Pharmacol. 2014;85(5):777–88. https://doi.org/10.1124/mol.113.091165.
Rothhammer V, Mascanfroni ID, Bunse L, Takenaka MC, Kenison JE, Mayo L, et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat Med. 2016;22:586–97. https://doi.org/10.1038/nm.4106.
Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22:598–605. https://doi.org/10.1038/nm.4102.
Schiering C, Wincent E, Metidji A, Iseppon A, Li Y, Potocnik AJ, et al. Feedback control of AHR signalling regulates intestinal immunity. Nature. 2017;542:242. https://doi.org/10.1038/nature21080Provides evidence that intestinal epithelial cells act as a gatekeeper to control the availability of dietary- or gut microbiota–derived AhR ligands.
Sasaki N, Sachs N, Wiebrands K, Ellenbroek SI, Fumagalli A, Lyubimova A, et al. Reg4+ deep crypt secretory cells function as epithelial niche for Lgr5+ stem cells in colon. Proc Natl Acad Sci U S A. 2016;113:E5399–407. https://doi.org/10.1073/pnas.1607327113.
Metidji A, Omenetti S, Crotta S, Li Y, Nye E, Ross E, et al. The environmental sensor AHR protects from inflammatory damage by maintaining intestinal stem cell homeostasis and barrier integrity. Immunity. 2018. https://doi.org/10.1016/j.immuni.2018.07.010Demonstrates that AhR signaling controls the maintenance and differentiation of intestinal stem cells by modulating Wnt/β-catenin signaling and ameliorates carcinogen-associated colon tumorigenesis.
Okino ST, Pookot D, Basak S, Dahiya R. Toxic and chemopreventive ligands preferentially activate distinct aryl hydrocarbon receptor pathways: implications for cancer prevention. Cancer Prev Res. 2009;2(3):251–6. https://doi.org/10.1158/1940-6207.Capr-08-0146.
Ahmad A, Ali S, Wang Z, Ali AS, Sethi S, Sakr WA, et al. 3,3′-Diindolylmethane enhances taxotere-induced growth inhibition of breast cancer cells through downregulation of FoxM1. Int J Cancer. 2011;129(7):1781–91. https://doi.org/10.1002/ijc.25839.
Hao H-X, Jiang X, Cong F. Control of Wnt receptor turnover by R-spondin-ZNRF3/RNF43 signaling module and its dysregulation in cancer. Cancers. 2016;8(6):54. https://doi.org/10.3390/cancers8060054.
Clevers H, Loh KM, Nusse R. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014;346(6205):1248012. https://doi.org/10.1126/science.1248012.
Diaz-Diaz CJ, Ronnekleiv-Kelly SM, Nukaya M, Geiger PG, Balbo S, Dator R, et al. The aryl hydrocarbon receptor is a repressor of inflammation-associated colorectal tumorigenesis in mouse. Ann Surg. 2016;264:429–36. https://doi.org/10.1097/SLA.0000000000001874.
Ema M, Ohe N, Suzuki M, Mimura J, Sogawa K, Ikawa S, et al. Dioxin binding activities of polymorphic forms of mouse and human arylhydrocarbon receptors. J Biol Chem. 1994;269(44):27337–43.
Flaveny CA, Murray IA, Perdew GH. Differential gene regulation by the human and mouse aryl hydrocarbon receptor. Toxicol Sci. 2010;114(2):217–25. https://doi.org/10.1093/toxsci/kfp308.
Goettel JA, Gandhi R, Kenison JE, Yeste A, Murugaiyan G, Sambanthamoorthy S, et al. AHR activation is protective against colitis driven by T cells in humanized mice. Cell Rep. 2016;17(5):1318–29. https://doi.org/10.1016/j.celrep.2016.09.082.
Natividad JM, Agus A, Planchais J, Lamas B, Jarry AC, Martin R, et al. Impaired aryl hydrocarbon receptor ligand production by the gut microbiota is a key factor in metabolic syndrome. Cell Metab. 2018;28(5):737-49 e4. https://doi.org/10.1016/j.cmet.2018.07.001Provides evidence that reduced gut microbiota–derived AhR ligands are observed in individuals with metabolic syndrome.
Boitano AE, Wang J, Romeo R, Bouchez LC, Parker AE, Sutton SE, et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010.
Harrill JA, Parks BB, Wauthier E, Rowlands JC, Reid LM, Thomas RS. Lineage-dependent effects of aryl hydrocarbon receptor agonists contribute to liver tumorigenesis. Hepatology. 2015;61(2):548–60. https://doi.org/10.1002/hep.27547.
Tofighi R, Wan Ibrahim WN, Rebellato P, Andersson PL, Uhlén P, Ceccatelli S. Non–dioxin-like polychlorinated biphenyls interfere with neuronal differentiation of embryonic neural stem cells. Toxicol Sci. 2011;124(1):192–201. https://doi.org/10.1093/toxsci/kfr221.
Biton M, Haber AL, Rogel N, Burgin G, Beyaz S, Schnell A, et al. T helper cell cytokines modulate intestinal stem cell renewal and differentiation. Cell. 2018;175(5):1307–20.e22. https://doi.org/10.1016/j.cell.2018.10.008.
Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E, et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453(7191):65–71. https://doi.org/10.1038/nature06880.
Kiss EA, Diefenbach A. Role of the aryl hydrocarbon receptor in controlling maintenance and functional programs of RORgammat(+) innate lymphoid cells and intraepithelial lymphocytes. Front Immunol. 2012;3:124. https://doi.org/10.3389/fimmu.2012.00124.
Li S, Bostick JW, Zhou L. Regulation of innate lymphoid cells by aryl hydrocarbon receptor. Front Immunol. 2018;8:1909. https://doi.org/10.3389/fimmu.2017.01909.
Li S, Bostick JW, Ye J, Qiu J, Zhang B, Urban JF Jr, et al. Aryl hydrocarbon receptor signaling cell intrinsically inhibits intestinal group 2 innate lymphoid cell function. Immunity. 2018;49(5):915–28.e5. https://doi.org/10.1016/j.immuni.2018.09.015Demonstrates that AhR signaling regulates the gut balance of ILC2 and ILC3 and supresses the number and functionality of ILC2 in a cell-intrinsic fashion.
Hayakawa Y, Wang TC. The tuft cell-ILC2 circuit integrates intestinal defense and homeostasis. Cell. 2018;174(2):251–3. https://doi.org/10.1016/j.cell.2018.06.037.
Yeste A, Mascanfroni ID, Nadeau M, Burns EJ, Tukpah A-M, Santiago A, et al. IL-21 induces IL-22 production in CD4+ T cells. Nat Commun. 2014;5:3753. https://doi.org/10.1038/ncomms4753.
Qiu J, Heller JJ, Guo X, Chen ZM, Fish K, Fu YX, et al. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36(1):92–104. https://doi.org/10.1016/j.immuni.2011.11.011.
Apetoh L, Quintana FJ, Pot C, Joller N, Xiao S, Kumar D, et al. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat Immunol. 2010;11(9):854–61. https://doi.org/10.1038/ni.1912.
Gutierrez-Vazquez C, Quintana FJ. Regulation of the immune response by the aryl hydrocarbon receptor. Immunity. 2018;48(1):19–33. https://doi.org/10.1016/j.immuni.2017.12.012Comprehensive review of current findings linking AhR signaling and host immune response.
Lindemans CA, Calafiore M, Mertelsmann AM, O'Connor MH, Dudakov JA, Jenq RR, et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015;528(7583):560–4. https://doi.org/10.1038/nature16460.
Zwarycz B, Gracz AD, Rivera KR, Williamson IA, Samsa LA, Starmer J, et al. IL22 inhibits epithelial stem cell expansion in an ileal organoid model. Cellular and Molecular Gastroenterology and Hepatology. 2019;7(1):1–17. https://doi.org/10.1016/j.jcmgh.2018.06.008.
Zha J-M, Li H-S, Lin Q, Kuo W-T, Jiang Z-H, Tsai P-Y, et al. Interleukin 22 expands transit-amplifying cells while depleting Lgr5+ stem cells via inhibition of Wnt and Notch signaling. Cellular and Molecular Gastroenterology and Hepatology. 2019;7(2):255–74. https://doi.org/10.1016/j.jcmgh.2018.09.006.
Jain U, Lai CW, Xiong S, Goodwin VM, Lu Q, Muegge BD, et al. Temporal regulation of the bacterial metabolite deoxycholate during colonic repair is critical for crypt regeneration. Cell Host Microbe. 2018;24(3):353–63.e5. https://doi.org/10.1016/j.chom.2018.07.019.
Roulis M, Kaklamanos A, Schernthanner M, Bielecki P, Zhao J, Kaffe E, et al. Paracrine orchestration of intestinal tumorigenesis by a mesenchymal niche. Nature. 2020;580(7804):524–9. https://doi.org/10.1038/s41586-020-2166-3.
Montrose DC, Nakanishi M, Murphy RC, Zarini S, McAleer JP, Vella AT, et al. The role of PGE2 in intestinal inflammation and tumorigenesis. Prostaglandins Other Lipid Mediat. 2015;116–117:26–36. https://doi.org/10.1016/j.prostaglandins.2014.10.002.
Takamura T, Harama D, Matsuoka S, Shimokawa N, Nakamura Y, Okumura K, et al. Activation of the aryl hydrocarbon receptor pathway may ameliorate dextran sodium sulfate-induced colitis in mice. Immunol Cell Biol. 2010;88(6):685–9. https://doi.org/10.1038/icb.2010.35.
Martey CA, Baglole CJ, Gasiewicz TA, Sime PJ, Phipps RP. The aryl hydrocarbon receptor is a regulator of cigarette smoke induction of the cyclooxygenase and prostaglandin pathways in human lung fibroblasts. Am J Phys Lung Cell Mol Phys. 2005;289(3):L391–L9. https://doi.org/10.1152/ajplung.00062.2005.
Wang D, Fu L, Sun H, Guo L, DuBois RN. Prostaglandin E2 promotes colorectal cancer stem cell expansion and metastasis in mice. Gastroenterology. 2015;149(7):1884–95.e4. https://doi.org/10.1053/j.gastro.2015.07.064.
Li Y, Soendergaard C, Bergenheim FH, Aronoff DM, Milne G, Riis LB, et al. COX-2-PGE2 signaling impairs intestinal epithelial regeneration and associates with TNF inhibitor responsiveness in ulcerative colitis. EBioMedicine. 2018;36:497–507. https://doi.org/10.1016/j.ebiom.2018.08.040.
Miyoshi H, VanDussen KL, Malvin NP, Ryu SH, Wang Y, Sonnek NM, et al. Prostaglandin E2 promotes intestinal repair through an adaptive cellular response of the epithelium. EMBO J. 2017;36(1):5–24. https://doi.org/10.15252/embj.201694660.
Garcia-Villatoro EL, DeLuca JAA, Callaway ES, Allred KF, Davidson LA, Hensel ME, et al. Effects of high-fat diet and intestinal aryl hydrocarbon receptor deletion on colon carcinogenesis. Am J Physiol Gastrointest Liver Physiol. 2020;318(3):G451–g63. https://doi.org/10.1152/ajpgi.00268.2019.
Jackstadt R, Sansom OJ. Mouse models of intestinal cancer. J Pathol. 2016;238(2):141–51. https://doi.org/10.1002/path.4645.
Washington K, Zemper AED. Apc-related models of intestinal neoplasia: a brief review for pathologists. Surgical and Experimental Pathology. 2019;2(1):11. https://doi.org/10.1186/s42047-019-0036-9.
Joseph R, Little P, Hayes DN, Lee MS. Characterization of the number and site of APC mutations in sporadic colorectal cancer. J Clin Onco. 2017;35(4_suppl):630. https://doi.org/10.1200/JCO.2017.35.4_suppl.630.
Yu AI, Zhao L, Eaton KA, Ho S, Chen J, Poe S, et al. Gut microbiota modulate CD8 T cell responses to influence colitis-associated tumorigenesis. Cell Rep. 2020;31(1):107471. https://doi.org/10.1016/j.celrep.2020.03.035.
Yoon K, Kim N. The effect of microbiota on colon carcinogenesis. J Cancer Prev. 2018;23(3):117–25. doi:https://doi.org/10.15430/JCP.2018.23.3.117.
Ikuta T, Kobayashi Y, Kitazawa M, Shiizaki K, Itano N, Noda T, et al. ASC-associated inflammation promotes cecal tumorigenesis in aryl hydrocarbon receptor-deficient mice. Carcinogenesis. 2013;34(7):1620–7. https://doi.org/10.1093/carcin/bgt083.
De Robertis M, Massi E, Poeta ML, Carotti S, Morini S, Cecchetelli L, et al. The AOM/DSS murine model for the study of colon carcinogenesis: from pathways to diagnosis and therapy studies. J Carcinog. 2011;10:9. https://doi.org/10.4103/1477-3163.78279.
Gronke K, Hernandez PP, Zimmermann J, Klose CSN, Kofoed-Branzk M, Guendel F, et al. Interleukin-22 protects intestinal stem cells against genotoxic stress. Nature. 2019;566(7743):249–53. https://doi.org/10.1038/s41586-019-0899-7Demonstrates that the production of IL22 by AhR signaling mainly in ILC3 protects intestinal stem cells from genotoxic insults, and reduces carcinogen-associated colon tumorigenesis.
Monteleone I, Rizzo A, Sarra M, Sica G, Sileri P, Biancone L, et al. Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology. 2011;141(1):237–48, 48 e1. https://doi.org/10.1053/j.gastro.2011.04.007.
Busbee PB, Menzel L, Alrafas HR, Dopkins N, Becker W, Miranda K, et al. Indole-3-carbinol prevents colitis and associated microbial dysbiosis in an IL-22–dependent manner. JCI Insight. 2020;5(1). https://doi.org/10.1172/jci.insight.127551Provides evidence that supplementation of the AhR ligand precursor indole-3-carbinol prevents colitis-associated microbial dysbiosis by induction of IL22.
Kawai S, Iijima H, Shinzaki S, Hiyama S, Yamaguchi T, Araki M, et al. Indigo naturalis ameliorates murine dextran sodium sulfate-induced colitis via aryl hydrocarbon receptor activation. J Gastroenterol. 2017;52(8):904–19. https://doi.org/10.1007/s00535-016-1292-z.
Bin P, Leng S, Cheng J, Dai Y, Huang C, Pan Z, et al. Association of aryl hydrocarbon receptor gene polymorphisms and urinary 1-hydroxypyrene in polycyclic aromatic hydrocarbon–exposed workers. Cancer Epidemiol Biomarkers Prevention. 2008;17(7):1702–8. https://doi.org/10.1158/1055-9965.Epi-07-2812.
Kovalova N, Manzan M, Crawford R, Kaminski N. Role of aryl hydrocarbon receptor polymorphisms on TCDD-mediated CYP1B1 induction and IgM suppression by human B cells. Toxicol Appl Pharmacol. 2016;309:15–23. https://doi.org/10.1016/j.taap.2016.08.011.
Wong JM, Okey AB, Harper PA. Human aryl hydrocarbon receptor polymorphisms that result in loss of CYP1A1 induction. Biochem Biophys Res Commun. 2001;288(4):990–6. https://doi.org/10.1006/bbrc.2001.5861.
Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47(9):979–86. https://doi.org/10.1038/ng.3359.
Gu A, Ji G, Long Y, Zhou Y, Shi X, Song L, et al. Assessment of an association between an aryl hydrocarbon receptor gene (AHR) polymorphism and risk of male infertility. Toxicol Sci. 2011;122(2):415–21. https://doi.org/10.1093/toxsci/kfr137.
Huang S, Shui X, He Y, Xue Y, Li J, Li G, et al. AhR expression and polymorphisms are associated with risk of coronary arterial disease in Chinese population. Sci Rep. 2015;5:8022. https://doi.org/10.1038/srep08022.
Rodgers GP, Collins FS. Precision Nutrition-the answer to “what to eat to stay healthy”. Jama. 2020;324:735–6. https://doi.org/10.1001/jama.2020.13601.
Acknowledgments
The illustrative figures were created using BioRender.com.
Funding
Funding was provided by Texas AgriLife Research, the Sid Kyle Chair Endowment, the Allen Endowed Chair in Nutrition & Chronic Disease Prevention, the Cancer Prevention Research Institute of Texas (RP160589), and the National Institutes of Health (R01-ES025713, R01-CA202697, R01-AT01282, R35-CA197707, and T32-CA090301).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Huajun Han, Arul Jayaraman, Stephen Safe, and Robert Chapkin declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Metabolism and Stem Cells
Rights and permissions
About this article
Cite this article
Han, H., Jayaraman, A., Safe, S. et al. Targeting the Aryl Hydrocarbon Receptor in Stem Cells to Improve the Use of Food as Medicine. Curr Stem Cell Rep 6, 109–118 (2020). https://doi.org/10.1007/s40778-020-00184-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40778-020-00184-0