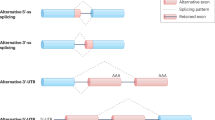
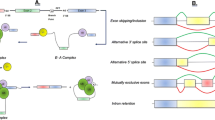
Abstract
Alzheimer’s disease (AD) is the most frequent neurodegenerative disorder in the elderly, occurring in approximately 20% of people older than 80. The molecular causes of AD are still poorly understood. However, recent studies have shown that Alternative Splicing (AS) is involved in the gene expression reprogramming associated with the functional changes observed in AD patients. In particular, mutations in cis-acting regulatory sequences as well as alterations in the activity and sub-cellular localization of trans-acting splicing factors and components of the spliceosome machinery are associated with splicing abnormalities in AD tissues, which may influence the onset and progression of the disease. In this review, we discuss the current molecular understanding of how alterations in the AS process contribute to AD pathogenesis. Finally, recent therapeutic approaches targeting aberrant AS regulation in AD are also reviewed.



Similar content being viewed by others
References
Albert MS, DeKosky ST, Dickson D et al (2011) The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement 7:270–279
Ittner LM, Götz J (2011) Amyloid-β and tau—a toxic pas de deux in Alzheimer’s disease. Nat Rev Neurosci 12:65–72
Braak H, Braak E (1991) Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol 82:239–259
Braak H, Alafuzoff I, Arzberger T et al (2006) Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol 112:389–404
Schellenberg GD, Bird TD, Wijsman EM et al (1992) Genetic linkage evidence for a familial Alzheimer’s disease locus on chromosome 14. Science 258:668–671
Vetrivel KS, Zhang YW, Xu H et al (2006) Pathological and physiological functions of presenilins. Mol Neurodegener 1:4
Alonso Vilatela ME, López-López M, Yescas-Gómez P (2012) Genetics of Alzheimer’s disease. Arch Med Res 43:622–631
Chabot B, Shkreta L (2016) Defective control of pre-messenger RNA splicing in human disease. J Cell Biol 212:13–27
Love JE, Hayden EJ, Rohn TT (2015) Alternative splicing in alzheimer’ s disease. J Park Dis Alzheimer’s Dis 2:6
Adusumalli S, Ngian ZK, Lin WQ et al (2019) Increased intron retention is a post-transcriptional signature associated with progressive aging and Alzheimer’s disease. Aging Cell 18:e12928
Braggin JE, Bucks SA, Course MM et al (2019) Alternative splicing in a presenilin 2 variant associated with Alzheimer disease. Ann Clin Transl Neurol 6:762–777
Han S, Miller J, Byun S et al (2019) Identification of exon skipping events associated with Alzheimer’s disease in the human hippocampus. BMC Med Genomics 12:13
Patel H, Hodges AK, Curtis C et al (2019) Transcriptomic analysis of probable asymptomatic and symptomatic alzheimer brains. Brain Behav Immun 80:644–656
Lee Y, Han S, Kim D et al (2018) Genetic variation affecting exon skipping contributes to brain structural atrophy in Alzheimer’s disease. AMIA Jt Summits Transl Sci Proc 2018:124–131
Tollervey JR, Wang Z, Hortobágyi T et al (2011) Analysis of alternative splicing associated with aging and neurodegeneration in the human brain. Genome Res 21:1572–1582
Treutlein B, Gokce O, Quake SR et al (2014) Cartography of neurexin alternative splicing mapped by single-molecule long-read mRNA sequencing. Proc Natl Acad Sci 111:E1291–E1299
Liu W, Wang F, Xu Q et al (2017) BCAS2 is involved in alternative mRNA splicing in spermatogonia and the transition to meiosis. Nat Commun 8:14182
Hannigan MM, Zagore LL, Licatalosi DD (2017) Ptbp2 controls an alternative splicing network required for cell communication during spermatogenesis. Cell Rep 19:2598–2612
Angiolini F, Belloni E, Giordano M et al (2019) A novel L1CAM isoform with angiogenic activity generated by NOVA2-mediated alternative splicing. Elife 8:e44305
Giampietro C, Deflorian G, Gallo S et al (2015) The alternative splicing factor Nova2 regulates vascular development and lumen formation. Nat Commun 6:8479
Yamamoto ML, Clark TA, Gee SL et al (2009) Alternative pre-mRNA splicing switches modulate gene expression in late erythropoiesis. Blood 113:3363–3370
Baralle FE, Giudice J (2017) Alternative splicing as a regulator of development and tissue identity. Nat Rev Mol Cell Biol 18:437–451
Martinez NM, Lynch KW (2013) Control of alternative splicing in immune responses: many regulators, many predictions, much still to learn. Immunol Rev 253:216–236
Ghigna C, Valacca C, Biamonti G (2008) Alternative splicing and tumor progression. Curr Genomics 9:556–570
Darnell RB (2013) RNA protein interaction in neurons. Annu Rev Neurosci 36:243–270
Raj B, Blencowe BJ (2015) Alternative splicing in the mammalian nervous system: recent insights into mechanisms and functional roles. Neuron 87:14–27
Lee S, Cieply B, Yang Y et al (2018) Esrp1-regulated splicing of Arhgef11 isoforms is required for epithelial tight junction integrity. Cell Rep 25:2417–2430.e5
Nagasaki H, Arita M, Nishizawa T et al (2005) Species-specific variation of alternative splicing and transcriptional initiation in six eukaryotes. Gene 364:53–62
Biamonti G, Maita L, Montecucco A (2018) The Krebs cycle connection: reciprocal influence between alternative splicing programs and cell metabolism. Front Oncol 8:408
Kornblihtt AR, Schor IE, Alló M et al (2013) Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol 14:153–165
Yeo G, Holste D, Kreiman G et al (2004) Variation in alternative splicing across human tissues. Genome Biol 5:R74
Dillman AA, Hauser DN, Gibbs JR et al (2013) MRNA expression, splicing and editing in the embryonic and adult mouse cerebral cortex. Nat Neurosci 16:499–506
Su CH, Dhananjaya D, Tarn WY (2018) Alternative splicing in neurogenesis and brain development. Front Mol Biosci 5:12
Licatalosi DD, Darnell RB (2006) Splicing regulation in neurologic disease. Neuron 52:93–101
Deschênes M, Chabot B (2017) The emerging role of alternative splicing in senescence and aging. Aging Cell 16:918–933
Grothe MJ, Sepulcre J, Gonzalez-Escamilla G et al (2018) Molecular properties underlying regional vulnerability to Alzheimer’s disease pathology. Brain 141:2755–2771
Twine NA, Janitz K, Wilkins MR et al (2011) Whole transcriptome sequencing reveals gene expression and splicing differences in brain regions affected by Alzheimer’s disease. PLoS ONE 6:e16266
Andersen K, Launer LJ, Dewey ME et al (1999) Gender differences in the incidence of AD and vascular dementia: the EURODEM studies. Neurology 53:1992–1997
Seshadri S, Wolf PA, Beiser A et al (1997) Lifetime risk of dementia and Alzheimer’s disease: the impact of mortality on risk estimates in the Framingham Study. Neurology 49:1498–1504
Fisher DW, Bennett DA, Dong H (2018) Sexual dimorphism in predisposition to Alzheimer’s disease. Neurobiol Aging 70:308–324
Dumitrescu L, Mayeda ER, Sharman K et al (2019) Sex differences in the genetic architecture of Alzheimer’s disease. Curr Genet Med Rep 7:13–21
Wang GS, Cooper TA (2007) Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet 8:749–761
De Strooper B, Iwatsubo T, Wolfe MS (2012) Presenilins and γ-secretase: structure, function, and role in Alzheimer disease. Cold Spring Harb Perspect Med 2:a006304
De Jonghe C, Cruts M, Rogaeva EA et al (1999) Aberrant splicing in the presenilin-1 intron 4 mutation causes presenile Alzheimer’s disease by increased Aβ42 secretion. Hum Mol Genet 8:1529–1540
Goode BL, Chau M, Denis PE et al (2002) Structural and functional differences between 3-Repeat and 4-Repeat tau isoforms. J Biol Chem 275:38182–38189
Espinoza M, De Silva R, Dickson DW et al (2008) Differential incorporation of tau isoforms in Alzheimer’s disease. J Alzheimer’s Dis 14:1–16
Buée L, Bussière T, Buée-Scherrer V et al (2000) Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res Brain Res Rev 33:95–130
Goedert M, Spillantini MG, Jakes R et al (1989) Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron 3:519–526
Ghetti B, Oblak AL, Boeve BF et al (2015) Invited review: frontotemporal dementia caused by microtubule-associated protein tau gene (MAPT) mutations: a chameleon for neuropathology and neuroimaging. Neuropathol Appl Neurobiol 41:24–46
Donahue CP, Muratore C, Wu JY et al (2006) Stabilization of the tau exon 10 stem loop alters pre-mRNA splicing. J Biol Chem 281:23302–23306
Allen M, Kachadoorian M, Quicksall Z et al (2014) Association of MAPT haplotypes with Alzheimer’s disease risk and MAPT brain gene expression levels. Alzheimer’s Res Ther 6:39
Valenca GT, Srivastava GP, Oliveira-Filho J et al (2016) The role of MAPT haplotype H2 and isoform 1N/4R in Parkinsonism of older adults. PLoS ONE 1:e0157452
Hoxha E, Lippiello P, Zurlo F et al (2018) The emerging role of altered cerebellar synaptic processing in Alzheimer’s disease. Front Aging Neurosci 10:396
Trabzuni D, Wray S, Vandrovcova J et al (2012) MAPT expression and splicing is differentially regulated by brain region: relation to genotype and implication for tauopathies. Hum Mol Genet 21:4094–4103
Trabzuni D, Ramasamy A, Imran S et al (2013) Widespread sex differences in gene expression and splicing in the adult human brain. Nat Commun 4:2771
Zou F, Gopalraj RK, Lok J et al (2008) Sex-dependent association of a common low-density lipoprotein receptor polymorphism with RNA splicing efficiency in the brain and Alzheimer’s disease. Hum Mol Genet 17:929–935
Lambert JC, Ibrahim-Verbaas CA, Harold D et al (2013) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 45:1452–1458
Raj T, Li YI, Wong G et al (2018) Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer’s disease susceptibility. Nat Genet 50:1584–1592
Raj T, Ryan KJ, Replogle JM et al (2014) CD33: increased inclusion of exon 2 implicates the Ig V-set domain in Alzheimer’s disease susceptibility. Hum Mol Genet 23:2729–2736
Li YI, van de Geijn B, Raj A et al (2016) RNA splicing is a primary link between genetic variation and disease. Science 352:600–604
Borreca A, Gironi K, Amadoro G et al (2016) Opposite dysregulation of fragile-X mental retardation protein and heteronuclear ribonucleoprotein C protein associates with enhanced APP translation in Alzheimer disease. Mol Neurobiol 53:3227–3234
Mueller SG, Weiner MW, Thal LJ et al (2005) The Alzheimer’s disease neuroimaging initiative. Neuroimaging Clin N Am 15:869–877
Kong LL, Miao D, Tan L et al (2018) Genome-wide association study identifies RBFOX1 locus influencing brain glucose metabolism. Ann Transl Med 6:436
Wamsley B, Jaglin XH, Favuzzi E et al (2018) Rbfox1 mediates cell-type-specific splicing in cortical interneurons. Neuron 100:846–859
Alkallas R, Fish L, Goodarzi H et al (2017) Inference of RNA decay rate from transcriptional profiling highlights the regulatory programs of Alzheimer’s disease. Nat Commun 8:909
Matsui T, Ingelsson M, Fukumoto H et al (2007) Expression of APP pathway mRNAs and proteins in Alzheimer’s disease. Brain Res 1161:116–123
Berson A, Barbash S, Shaltiel G et al (2012) Cholinergic-associated loss of hnRNP-A/B in Alzheimer’s disease impairs cortical splicing and cognitive function in mice. EMBO Mol Med 4:730–742
Ferreira-Vieira TH, Guimaraes IM, Silva FR et al (2016) Alzheimer’s disease: targeting the cholinergic system. Curr Neuropharmacol 14:101–115
Kolisnyk B, Al-Onaizi M, Soreq L et al (2017) Cholinergic surveillance over hippocampal RNA metabolism and Alzheimer’s-like pathology. Cereb Cortex 27:3553–3567
Lu J, Shu R, Zhu Y (2018) Dysregulation and dislocation of SFPQ disturbed DNA organization in Alzheimer’s disease and frontotemporal dementia. J Alzheimer’s Dis 61:1311–1321
Ke YD, Dramiga J, Schütz U et al (2012) Tau-mediated nuclear depletion and cytoplasmic accumulation of SFPQ in Alzheimer’s and Pick’s disease. PLoS ONE 7:e35678
Ray P, Kar A, Fushimi K et al (2011) PSF suppresses tau exon 10 inclusion by interacting with a stem-loop structure downstream of exon 10. J Mol Neurosci 45:453–466
Buratti E, Baralle FE (2008) Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front Biosci 13:867–878
Neumann M, Sampathu DM, Kwong LK et al (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314:130–133
Amador-Ortiz C, Lin WL, Ahmed Z et al (2007) TDP-43 immunoreactivity in hippocampal sclerosis and Alzheimer’s disease. Ann Neurol 61:435–445
Wang G, Yang H, Yan S et al (2015) Cytoplasmic mislocalization of RNA splicing factors and aberrant neuronal gene splicing in TDP-43 transgenic pig brain. Mol Neurodegener 10:42
Bai B, Hales CM, Chen PC et al (2013) U1 small nuclear ribonucleoprotein complex and RNA splicing alterations in Alzheimer’s disease. Proc Natl Acad Sci 110:16562–16567
Hales CM, Dammer EB, Diner I et al (2014) Aggregates of small nuclear ribonucleic acids (snRNAs) in Alzheimer’s disease. Brain Pathol 24:344–351
Bai B (2018) U1 snRNP alteration and neuronal cell cycle reentry in Alzheimer disease. Front Aging Neurosci 10:75
Yang Y, Geldmacher DS, Herrup K (2011) DNA replication precedes neuronal cell death in Alzheimer’s disease. J Neurosci 21:2661–2668
Kroemer G, El-Deiry WS, Golstein P et al (2009) Classification of cell death: recommendations of the nomenclature committee on cell death 2009. Cell Death Differ 12:1463–1467
Heppner FL, Ransohoff RM, Becher B (2015) Immune attack: the role of inflammation in Alzheimer disease. Nat Rev Neurosci 16:358–372
Ule J, Ule A, Spencer J et al (2005) Nova regulates brain-specific splicing to shape the synapse. Nat Genet 37:844–852
Ruggiu M, Herbst R, Kim N et al (2009) Rescuing Z+ agrin splicing in Nova null mice restores synapse formation and unmasks a physiologic defect in motor neuron firing. Proc Natl Acad Sci 106:3513–3518
Iruela-Arispe ML, Davis GE (2009) Cellular and molecular mechanisms of vascular lumen formation. Dev Cell 16:222–231
Solecki DJ, Govek EE, Tomoda T et al (2006) Neuronal polarity in CNS development. Genes Dev 20:2639–2647
Segura I, De Smet F, Hohensinner PJ et al (2009) The neurovascular link in health and disease: an update. Trends Mol Med 15:439–451
Quaegebeur A, Lange C, Carmeliet P (2011) The neurovascular link in health and disease: molecular mechanisms and therapeutic implications. Neuron 71:406–424
Chang JL, Hinrich AJ, Roman B et al (2018) Targeting amyloid-β precursor protein, APP, splicing with antisense oligonucleotides reduces toxic amyloid-β production. Mol Ther 26:1539–1551
Hinrich AJ, Jodelka FM, Chang JL et al (2016) Therapeutic correction of ApoER2 splicing in Alzheimer’s disease mice using antisense oligonucleotides. EMBO Mol Med 8:328–345
Wasser CR, Herz J (2016) Splicing therapeutics for Alzheimer’s disease. EMBO Mol Med 8:308–310
Schoch KM, DeVos SL, Miller RL et al (2016) Increased 4R-tau induces pathological changes in a human-tau mouse model. Neuron 90:941–947
Peacey E, Rodriguez L, Liu Y et al (2012) Targeting a pre-mRNA structure with bipartite antisense molecules modulates tau alternative splicing. Nucleic Acids Res 40:9836–9849
Rodriguez-Martin T, Garcia-Blanco MA, Mansfield SG et al (2005) Reprogramming of tau alternative splicing by spliceosome-mediated RNA trans-splicing: implications for tauopathies. Proc Natl Acad Sci 102:15659–15664
Avale ME, Rodríguez-Martín T, Gallo JM (2013) Trans-splicing correction of tau isoform imbalance in a mouse model of tau mis-splicing. Hum Mol Genet 22:2603–2611
Espíndola SL, Damianich A, Alvarez RJ et al (2018) Modulation of tau isoforms imbalance precludes tau pathology and cognitive decline in a mouse model of tauopathy. Cell Rep 23:709–715
Tanaka H, Kondo K, Chen X et al (2018) The intellectual disability gene PQBP1 rescues Alzheimer’s disease pathology. Mol Psychiatry 23:2090–2110
György B, Lööv C, Zaborowski MP et al (2018) CRISPR/Cas9 mediated disruption of the Swedish APP allele as a therapeutic approach for early-onset Alzheimer’s disease. Mol Ther Nucl Acids 11:429–440
Konermann S, Lotfy P, Brideau NJ et al (2018) Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 173:665–676
Radde R, Duma C, Goedert M et al (2008) The value of incomplete mouse models of Alzheimer’s disease. Eur J Nucl Med Mol Imaging 35:S70–S74
Blencowe BJ (2006) Alternative splicing: new insights from global analyses. Cell 126:37–47
Acknowledgements
We apologize to the authors of many interesting and important papers that have not been cited in this review.
Funding
This work was supported by the National Research Council of Italy (CNR), Research Project “Aging: molecular and technological innovations for improving the health of the elderly population” (Prot. MIUR 2867 25.11.2011) and AMANDA project Accordo Quadro Regione Lombardia–CNR to G.B. E.B. is supported by AIRC - FIRC ITALY postdoctoral fellowship; D.P. is awarded with a Arturo Falaschi - Fondazione Adriano Buzzati-Traverso (FAB-T) postdoctoral fellowship.
Author information
Authors and Affiliations
Contributions
All authors contributed to all parts of the present review including papers selection, information synthesis, and paper drafting/editing. All authors approved and verified the final version.
Corresponding author
Ethics declarations
Conflict of interest
C.G. is a consultant for Gene Tools. Funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Statement of human and animal rights
This review does not include studies with human or animal samples performed by any of the authors.
Informed consent
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Biamonti, G., Amato, A., Belloni, E. et al. Alternative splicing in Alzheimer’s disease. Aging Clin Exp Res 33, 747–758 (2021). https://doi.org/10.1007/s40520-019-01360-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40520-019-01360-x