Abstract
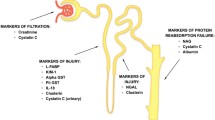
Early detection of graft injury after kidney transplantation is key to maintaining long-term good graft function. Graft injury could be due to a multitude of factors including ischaemia reperfusion injury, cell or antibody-mediated rejection, progressive interstitial fibrosis and tubular atrophy, infections and toxicity from the immunosuppressive drugs themselves. The current gold standard for assessing renal graft dysfunction is renal biopsy. However, biopsy is usually late when triggered by a change in serum creatinine and of limited utility in diagnosis of early injury when histological changes are equivocal. Therefore, there is a need for timely, objective and non-invasive diagnostic techniques with good early predictive value to determine graft injury and provide precision in titrating immunosuppression. We review potential novel plasma and urine biomarkers that offer sensitive new strategies for early detection and provide major insights into mechanisms of graft injury. This is a rapidly expanding field, but it is likely that a combination of biomarkers will be required to provide adequate sensitivity and specificity for detecting graft injury.



Similar content being viewed by others
References
Hill NR, Fatoba ST, Oke JL, Hirst JA, O’Callaghan CA, Lasserson DS, et al. Global prevalence of chronic kidney disease—a systematic review and meta-analysis. PLoS One. 2016;11(7):e0158765. https://doi.org/10.1371/journal.pone.0158765.
Wang JH, Skeans MA, Israni AK. Current status of kidney transplant outcomes: dying to survive. Adv Chronic Kidney Dis. 2016;23(5):281–6. https://doi.org/10.1053/j.ackd.2016.07.001.
Lechler RI, Sykes M, Thomson AW, Turka LA. Organ transplantation–how much of the promise has been realized? Nat Med. 2005;11(6):605–13. https://doi.org/10.1038/nm1251.
Salvadori M, Rosso G, Bertoni E. Update on ischemia-reperfusion injury in kidney transplantation: pathogenesis and treatment. World J Transplant. 2015;5(2):52–67. https://doi.org/10.5500/wjt.v5.i2.52.
Wu WK, Famure O, Li Y, Kim SJ. Delayed graft function and the risk of acute rejection in the modern era of kidney transplantation. Kidney Int. 2015;88(4):851–8. https://doi.org/10.1038/ki.2015.190.
Salvadori M, Tsalouchos A. Biomarkers in renal transplantation: an updated review. World J Transplant. 2017;7(3):161–78. https://doi.org/10.5500/wjt.v7.i3.161.
Bohra R, Klepacki J, Klawitter J, Klawitter J, Thurman JM, Christians U. Proteomics and metabolomics in renal transplantation-quo vadis? Transpl Int. 2013;26(3):225–41. https://doi.org/10.1111/tri.12003.
Mohammadpour N, Elyasi S, Vahdati N, Mohammadpour AH, Shamsara J. A review on therapeutic drug monitoring of immunosuppressant drugs. Iran J Basic Med Sci. 2011;14(6):485–98.
Fernando M, Peake PW, Endre ZH. Biomarkers of calcineurin inhibitor nephrotoxicity in transplantation. Biomark Med. 2014;8(10):1247–62. https://doi.org/10.2217/bmm.14.86.
Gwinner W, Metzger J, Husi H, Marx D. Proteomics for rejection diagnosis in renal transplant patients: where are we now? World J Transplant. 2016;6(1):28–41. https://doi.org/10.5500/wjt.v6.i1.28.
Haas M, Loupy A, Lefaucheur C, Roufosse C, Glotz D, Seron D, et al. The Banff 2017 Kidney Meeting Report: revised diagnostic criteria for chronic active T cell-mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. Am J Transplant. 2018;18(2):293–307. https://doi.org/10.1111/ajt.14625.
Shipkova M, Wieland E. Editorial: immune monitoring in solid organ transplantation. Clin Biochem. 2016;49(4):317–9. https://doi.org/10.1016/j.clinbiochem.2016.01.005.
Hollis E, Shehata M, Khalifa F, El-Ghar MA, El-Diasty T, El-Baza A. Towards non-invasive diagnostic techniques for early detection of acute renal transplant rejection: a review. Egypt J Radiol Nucl Med. 2017. https://doi.org/10.1016/j.ejrnm.2016.11.005.
Succar L, Pianta TJ, Davidson T, Pickering JW, Endre ZH. Subclinical chronic kidney disease modifies the diagnosis of experimental acute kidney injury. Kidney Int. 2017;92(3):680–92. https://doi.org/10.1016/j.kint.2017.02.030.
Myers GL, Miller WG, Coresh J, Fleming J, Greenberg N, Greene T, et al. Recommendations for improving serum creatinine measurement: a report from the Laboratory Working Group of the National Kidney Disease Education Program. Clin Chem. 2006;52(1):5–18. https://doi.org/10.1373/clinchem.2005.0525144.
Solez K, Racusen LC. The Banff classification revisited. Kidney Int. 2013;83(2):201–6. https://doi.org/10.1038/ki.2012.395.
Meersch M, Schmidt C, Hoffmeier A, Van Aken H, Wempe C, Gerss J, et al. Prevention of cardiac surgery-associated AKI by implementing the KDIGO guidelines in high risk patients identified by biomarkers: the PrevAKI randomized controlled trial. Intensive Care Med. 2017;43(11):1551–61. https://doi.org/10.1007/s00134-016-4670-3.
Pianta TJ, Peake PW, Pickering JW, Kelleher M, Buckley NA, Endre ZH. Evaluation of biomarkers of cell cycle arrest and inflammation in prediction of dialysis or recovery after kidney transplantation. Transpl Int. 2015;28(12):1392–404. https://doi.org/10.1111/tri.12636.
Christians U, Klawitter J, Klawitter J. Biomarkers in transplantation—proteomics and metabolomics. Ther Drug Monit. 2016;38(Suppl 1):S70–4. https://doi.org/10.1097/ftd.0000000000000243.
Pianta TJ, Peake PW, Pickering JW, Kelleher M, Buckley NA, Endre ZH. Clusterin in kidney transplantation: novel biomarkers versus serum creatinine for early prediction of delayed graft function. Transplantation. 2015;99(1):171–9. https://doi.org/10.1097/tp.0000000000000256.
Pianta TJ, Succar L, Davidson T, Buckley NA, Endre ZH. Monitoring treatment of acute kidney injury with damage biomarkers. Toxicol Lett. 2017;268:63–70. https://doi.org/10.1016/j.toxlet.2017.01.001.
Vilar E, Varagunam M, Yaqoob MM, Raftery M, Thuraisingham R. Creatinine reduction ratio: a useful marker to identify medium and high-risk renal transplants. Transplantation. 2010;89(1):97–103. https://doi.org/10.1097/TP.0b013e3181be3dd1.
Yarlagadda SG, Coca SG, Formica RN Jr, Poggio ED, Parikh CR. Association between delayed graft function and allograft and patient survival: a systematic review and meta-analysis. Nephrol Dial Transplant. 2009;24(3):1039–47. https://doi.org/10.1093/ndt/gfn667.
Moers C, Smits JM, Maathuis MH, Treckmann J, van Gelder F, Napieralski BP, et al. Machine perfusion or cold storage in deceased-donor kidney transplantation. N Engl J Med. 2009;360(1):7–19. https://doi.org/10.1056/NEJMoa0802289.
Ronco C, Legrand M, Goldstein SL, Hur M, Tran N, Howell EC, et al. Neutrophil gelatinase-associated lipocalin: ready for routine clinical use? An international perspective. Blood Purif. 2014;37(4):271–85. https://doi.org/10.1159/000360689.
Devarajan P. Neutrophil gelatinase-associated lipocalin (NGAL): a new marker of kidney disease. Scand J Clin Lab Investig Suppl. 2008;241:89–94. https://doi.org/10.1080/00365510802150158.
Fonseca I, Oliveira JC, Almeida M, Cruz M, Malho A, Martins LS, et al. Neutrophil gelatinase-associated lipocalin in kidney transplantation is an early marker of graft dysfunction and is associated with 1-year renal function. J Transplant. 2013;2013:650123. https://doi.org/10.1155/2013/650123.
Hall IE, Yarlagadda SG, Coca SG, Wang Z, Doshi M, Devarajan P, et al. IL-18 and urinary NGAL predict dialysis and graft recovery after kidney transplantation. J Am Soc Nephrol. 2010;21(1):189–97. https://doi.org/10.1681/asn.2009030264.
Pezeshgi A, Azar SA, Ghasemi H, Kamali K, Esmaeilzadeh A, Hajsalimi B, et al. Role of plasma neutrophil gelatinase-associated lipocalin as an emerging biomarker of acute renal failure following kidney transplantation and its correlation with plasma creatinine. J Ren Inj Prev. 2016;5(2):98–103. https://doi.org/10.15171/jrip.2016.21.
Cantaluppi V, Dellepiane S, Tamagnone M, Medica D, Figliolini F, Messina M, et al. Neutrophil gelatinase associated lipocalin is an early and accurate biomarker of graft function and tissue regeneration in kidney transplantation from extended criteria donors. PLoS One. 2015;10(6):e0129279. https://doi.org/10.1371/journal.pone.0129279.
Peake PW, Pianta TJ, Succar L, Fernando M, Pugh DJ, McNamara K, et al. A comparison of the ability of levels of urinary biomarker proteins and exosomal mRNA to predict outcomes after renal transplantation. PLoS One. 2014;9(2):e98644. https://doi.org/10.1371/journal.pone.0098644.
Susal C, Wettstein D, Dohler B, Morath C, Ruhenstroth A, Scherer S, et al. Association of kidney graft loss with de novo produced donor-specific and non-donor-specific HLA antibodies detected by single antigen testing. Transplantation. 2015;99(9):1976–80. https://doi.org/10.1097/tp.0000000000000672.
Yamamoto T, Watarai Y, Takeda A, Tsujita M, Hiramitsu T, Goto N, et al. De novo anti-HLA DSA characteristics and subclinical antibody-mediated kidney allograft injury. Transplantation. 2016;100(10):2194–202. https://doi.org/10.1097/tp.0000000000001012.
Wiebe C, Gibson IW, Blydt-Hansen TD, Karpinski M, Ho J, Storsley LJ, et al. Evolution and clinical pathologic correlations of de novo donor-specific HLA antibody post kidney transplant. Am J Transplant. 2012;12(5):1157–67. https://doi.org/10.1111/j.1600-6143.2012.04013.x.
Ginevri F, Nocera A, Comoli P, Innocente A, Cioni M, Parodi A, et al. Posttransplant de novo donor-specific HLA antibodies identify pediatric kidney recipients at risk for late antibody-mediated rejection. Am J Transplant. 2012;12(12):3355–62. https://doi.org/10.1111/j.1600-6143.2012.04251.x.
Wiebe C, Gibson IW, Blydt-Hansen TD, Pochinco D, Birk PE, Ho J, et al. Rates and determinants of progression to graft failure in kidney allograft recipients with de novo donor-specific antibody. Am J Transplant. 2015;15(11):2921–30. https://doi.org/10.1111/ajt.13347.
Charnaya O, Tuchman S, Moudgil A. Results of early treatment for de novo donor-specific antibodies in pediatric kidney transplant recipients in a cross-sectional and longitudinal cohort. Pediatr Transplant. 2018. https://doi.org/10.1111/petr.13108.
Billing H, Rieger S, Susal C, Waldherr R, Opelz G, Wuhl E, et al. IVIG and rituximab for treatment of chronic antibody-mediated rejection: a prospective study in paediatric renal transplantation with a 2-year follow-up. Transpl Int. 2012;25(11):1165–73. https://doi.org/10.1111/j.1432-2277.2012.01544.x.
Orandi BJ, Chow EH, Hsu A, Gupta N, Van Arendonk KJ, Garonzik-Wang JM, et al. Quantifying renal allograft loss following early antibody-mediated rejection. Am J Transplant. 2015;15(2):489–98. https://doi.org/10.1111/ajt.12982.
Cooper JE, Gralla J, Klem P, Chan L, Wiseman AC. High dose intravenous immunoglobulin therapy for donor-specific antibodies in kidney transplant recipients with acute and chronic graft dysfunction. Transplantation. 2014;97(12):1253–9. https://doi.org/10.1097/01.Tp.0000443226.74584.03.
Aubert O, Loupy A, Hidalgo L, van Huyen JPD, Higgins S, Viglietti D, et al. Antibody-mediated rejection due to preexisting versus de novo donor-specific antibodies in kidney allograft recipients. J Am Soc Nephrol. 2017;28(6):1912–23. https://doi.org/10.1681/asn.2016070797.
Menon MC, Murphy B, Heeger PS. Moving biomarkers toward clinical implementation in kidney transplantation. J Am Soc Nephrol. 2017;28(3):735–47. https://doi.org/10.1681/asn.2016080858.
Mueller TF, Reeve J, Jhangri GS, Mengel M, Jacaj Z, Cairo L, et al. The transcriptome of the implant biopsy identifies donor kidneys at increased risk of delayed graft function. Am J Transplant. 2008;8(1):78–85. https://doi.org/10.1111/j.1600-6143.2007.02032.x.
Kreepala C, Famulski KS, Chang J, Halloran PF. Comparing molecular assessment of implantation biopsies with histologic and demographic risk assessment. Am J Transplant. 2013;13(2):415–26. https://doi.org/10.1111/ajt.12043.
Khatri P, Roedder S, Kimura N, De Vusser K, Morgan AA, Gong Y, et al. A common rejection module (CRM) for acute rejection across multiple organs identifies novel therapeutics for organ transplantation. J Exp Med. 2013;210(11):2205–21. https://doi.org/10.1084/jem.20122709.
Sigdel TK, Bestard O, Tran TQ, Hsieh SC, Roedder S, Damm I, et al. A computational gene expression score for predicting immune injury in renal allografts. PLoS One. 2015;10(9):e0138133. https://doi.org/10.1371/journal.pone.0138133.
Reeve J, Sellares J, Mengel M, Sis B, Skene A, Hidalgo L, et al. Molecular diagnosis of T cell-mediated rejection in human kidney transplant biopsies. Am J Transplant. 2013;13(3):645–55. https://doi.org/10.1111/ajt.12079.
Limmathurotsakul D, Turner EL, Wuthiekanun V, Thaipadungpanit J, Suputtamongkol Y, Chierakul W, et al. Fool’s gold: why imperfect reference tests are undermining the evaluation of novel diagnostics: a reevaluation of 5 diagnostic tests for leptospirosis. Clin Infect Dis. 2012;55(3):322–31. https://doi.org/10.1093/cid/cis403.
Halloran PF, Pereira AB, Chang J, Matas A, Picton M, De Freitas D, et al. Microarray diagnosis of antibody-mediated rejection in kidney transplant biopsies: an international prospective study (INTERCOM). Am J Transplant. 2013;13(11):2865–74. https://doi.org/10.1111/ajt.12465.
Loupy A, Lefaucheur C, Vernerey D, Chang J, Hidalgo LG, Beuscart T, et al. Molecular microscope strategy to improve risk stratification in early antibody-mediated kidney allograft rejection. J Am Soc Nephrol. 2014;25(10):2267–77. https://doi.org/10.1681/asn.2013111149.
Traitanon O, Poggio ED, Fairchild RL. Molecular monitoring of alloimmune-mediated injury in kidney transplant patients. Curr Opin Nephrol Hypertens. 2014;23(6):625–30. https://doi.org/10.1097/mnh.0000000000000064.
Mas VR, Dumur CI, Scian MJ, Gehrau RC, Maluf DG. MicroRNAs as biomarkers in solid organ transplantation. Am J Transplant. 2013;13(1):11–9. https://doi.org/10.1111/j.1600-6143.2012.04313.x.
Danger R, Paul C, Giral M, Lavault A, Foucher Y, Degauque N, et al. Expression of miR-142-5p in peripheral blood mononuclear cells from renal transplant patients with chronic antibody-mediated rejection. PLoS One. 2013;8(4):e60702. https://doi.org/10.1371/journal.pone.0060702.
Wilflingseder J, Reindl-Schwaighofer R, Sunzenauer J, Kainz A, Heinzel A, Mayer B, et al. MicroRNAs in kidney transplantation. Nephrol Dial Transplant. 2015;30(6):910–7. https://doi.org/10.1093/ndt/gfu280.
Bloom RD, Bromberg JS, Poggio ED, Bunnapradist S, Langone AJ, Sood P, et al. Cell-free DNA and active rejection in kidney allografts. J Am Soc Nephrol. 2017;28(7):2221–32. https://doi.org/10.1681/asn.2016091034.
Adamek M, Opelz G, Klein K, Morath C, Tran TH. A fast and simple method for detecting and quantifying donor-derived cell-free DNA in sera of solid organ transplant recipients as a biomarker for graft function. Clin Chem Lab Med. 2016;54(7):1147–55. https://doi.org/10.1515/cclm-2015-0622.
Garcia Moreira V, Prieto Garcia B, Baltar Martin JM, Ortega Suarez F, Alvarez FV. Cell-free DNA as a noninvasive acute rejection marker in renal transplantation. Clin Chem. 2009;55(11):1958–66. https://doi.org/10.1373/clinchem.2009.129072.
Sigdel TK, Sarwal MM. Cell-free DNA as a measure of transplant injury. Clin Transpl. 2012;201–5.
Celec P, Vlkova B, Laukova L, Babickova J, Boor P. Cell-free DNA: the role in pathophysiology and as a biomarker in kidney diseases. Expert Rev Mol Med. 2018;20:e1. https://doi.org/10.1017/erm.2017.12.
Beck J, Bierau S, Balzer S, Andag R, Kanzow P, Schmitz J, et al. Digital droplet PCR for rapid quantification of donor DNA in the circulation of transplant recipients as a potential universal biomarker of graft injury. Clin Chem. 2013;59(12):1732–41. https://doi.org/10.1373/clinchem.2013.210328.
Schütz E, Fischer A, Beck J, Harden M, Koch M, Wuensch T, et al. Graft-derived cell-free DNA, a noninvasive early rejection and graft damage marker in liver transplantation: a prospective, observational, multicenter cohort study. PLoS Med. 2017;14(4):e1002286. https://doi.org/10.1371/journal.pmed.1002286.
Knight SR, Thorne A, Lo Faro ML. Donor-specific cell-free DNA as a biomarker in solid organ transplantation. A systematic review. Transplantation. 2019;103(2):273–83. https://doi.org/10.1097/tp.0000000000002482.
Lee H, Park Y-M, We Y-M, Han DJ, Seo J-W, Moon H, et al. Evaluation of digital PCR as a technique for monitoring acute rejection in kidney transplantation. Genom Inform. 2017;15(1):2–10. https://doi.org/10.5808/GI.2017.15.1.2.
Grskovic M, Hiller DJ, Eubank LA, Sninsky JJ, Christopherson C, Collins JP, et al. Validation of a clinical-grade assay to measure donor-derived cell-free DNA in solid organ transplant recipients. J Mol Diagn. 2016;18(6):890–902. https://doi.org/10.1016/j.jmoldx.2016.07.003.
Gielis EM, Beirnaert C, Dendooven A, Meysman P, Laukens K, De Schrijver J, et al. Plasma donor-derived cell-free DNA kinetics after kidney transplantation using a single tube multiplex PCR assay. PLoS One. 2018;13(12):e0208207. https://doi.org/10.1371/journal.pone.0208207.
Stoltz D, Brubaker A, Grskovic M, Woodward R, Gallo A. Donor-derived cell-free DNA predicts biopsy-proven acute cellular rejection in pediatric kidney transplant recipients [abstract no. 483]. 2017 American Transplant Congress; 29 Apr–3 May. Chicago; 2017.
Sigdel TK, Vitalone MJ, Tran TQ, Dai H, Hsieh S-C, Salvatierra O, et al. A rapid noninvasive assay for the detection of renal transplant injury. Transplantation. 2013;96(1):97–101. https://doi.org/10.1097/TP.0b013e318295ee5a.
Sigdel T, Tran T, Hsieh S-C, Sarwal M. Urinary cell-free dna is a sensitive marker of early renal transplant injury [abstract no. B272]. 2015 American Transplant Congress; 2–6 May; Philadelphia; 2015.
Beck J, Kanzow P, Schmitz J, Kollmar O, Oellerich M, Schütz E. Absolute quantification of graft derived cell-free DNA (GcfDNA) early after liver transplantation (LTx) using droplet digital PCR. Clin Chem. 2014;60(Suppl):S194–5.
Whitlam JB, Ling L, Skene A, Kanellis J, Ierino FL, Slater HR, et al. Diagnostic application of kidney allograft-derived absolute cell-free DNA levels during transplant dysfunction. Am J Transplant. 2018. https://doi.org/10.1111/ajt.15142 (Epub 2018 Oct 12).
Bromberg JS, Brennan DC, Poggio E, Bunnapradist S, Langone A, Sood P, et al. Biological variation of donor-derived cell-free dna in renal transplant recipients: clinical implications. J Appl Lab Med. 2017;2(3):309–21. https://doi.org/10.1373/jalm.2016.022731.
Sigdel TK, Archila FA, Constantin T, Prins SA, Liberto J, Damm I, et al. Optimizing detection of kidney transplant injury by assessment of donor-derived cell-free DNA via massively multiplex PCR. J Clin Med. 2018. https://doi.org/10.3390/jcm8010019.
Goh SK, Muralidharan V, Christophi C, Do H, Dobrovic A. Probe-free digital PCR quantitative methodology to measure donor-specific cell-free DNA after solid-organ transplantation. Clin Chem. 2017;63(3):742–50. https://doi.org/10.1373/clinchem.2016.264838.
Wishart DS. Metabolomics: a complementary tool in renal transplantation. Contrib Nephrol. 2008;160:76–87. https://doi.org/10.1159/000125935.
Naesens M, Sarwal MM. Molecular diagnostics in transplantation. Nat Rev Nephrol. 2010;6:614–28. https://doi.org/10.1038/nrneph.2010.113.
Nasr M, Sigdel T, Sarwal M. Advances in diagnostics for transplant rejection. Expert Rev Mol Diagn. 2016;16(10):1121–32. https://doi.org/10.1080/14737159.2016.1239530.
Blydt-Hansen TD, Sharma A, Gibson IW, Wishart DS, Mandal R, Ho J, et al. Urinary metabolomics for noninvasive detection of antibody-mediated rejection in children after kidney transplantation. Transplantation. 2017;101(10):2553–61. https://doi.org/10.1097/tp.0000000000001662.
Mincham CM, Gibson IW, Sharma A, Wiebe C, Mandal R, Rush D, et al. Evolution of renal function and urinary biomarker indicators of inflammation on serial kidney biopsies in pediatric kidney transplant recipients with and without rejection. Pediatr Transplant. 2018;22(5):e13202. https://doi.org/10.1111/petr.13202.
Foxall PJ, Mellotte GJ, Bending MR, Lindon JC, Nicholson JK. NMR spectroscopy as a novel approach to the monitoring of renal transplant function. Kidney Int. 1993;43(1):234–45.
Klawitter J, Haschke M, Kahle C, Dingmann C, Klawitter J, Leibfritz D, et al. Toxicodynamic effects of ciclosporin are reflected by metabolite profiles in the urine of healthy individuals after a single dose. Br J Clin Pharmacol. 2010;70(2):241–51. https://doi.org/10.1111/j.1365-2125.2010.03689.x.
Klepacki J, Klawitter J, Klawitter J, Thurman JM, Christians U. A high-performance liquid chromatography-tandem mass spectrometry-based targeted metabolomics kidney dysfunction marker panel in human urine. Clin Chim Acta. 2015;446:43–53. https://doi.org/10.1016/j.cca.2015.04.005.
Endre ZH, Pickering JW, Storer MK, Hu WP, Moorhead KT, Allardyce R, et al. Breath ammonia and trimethylamine allow real-time monitoring of haemodialysis efficacy. Physiol Meas. 2011;32(1):115–30. https://doi.org/10.1088/0967-3334/32/1/008.
Stubbs JR, House JA, Ocque AJ, Zhang S, Johnson C, Kimber C, et al. Serum trimethylamine-N-oxide is elevated in CKD and correlates with coronary atherosclerosis burden. J Am Soc Nephrol. 2016;27(1):305–13. https://doi.org/10.1681/asn.2014111063.
Bonneau E, Tetreault N, Robitaille R, Boucher A, De Guire V. Metabolomics: perspectives on potential biomarkers in organ transplantation and immunosuppressant toxicity. Clin Biochem. 2016;49(4–5):377–84. https://doi.org/10.1016/j.clinbiochem.2016.01.006.
Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368(17):1575–84. https://doi.org/10.1056/NEJMoa1109400.
Kim KB, Yang JY, Kwack SJ, Park KL, Kim HS, Ryu DH, et al. Toxicometabolomics of urinary biomarkers for human gastric cancer in a mouse model. J Toxicol Environ Health A. 2010;73(21–22):1420–30. https://doi.org/10.1080/15287394.2010.511545.
Calderisi M, Vivi A, Mlynarz P, Tassin M, Banasik M, Dawiskiba T, et al. Using metabolomics to monitor kidney transplantation patients by means of clustering to spot anomalous patient behavior. Transplant Proc. 2013;45(4):1511–5. https://doi.org/10.1016/j.transproceed.2013.02.049.
Karpievitch YV, Polpitiya AD, Anderson GA, Smith RD, Dabney AR. Liquid chromatography mass spectrometry-based proteomics: biological and technological aspects. Ann Appl Stat. 2010;4(4):1797–823. https://doi.org/10.1214/10-AOAS341.
Anglicheau D, Naesens M, Essig M, Gwinner W, Marquet P. Establishing biomarkers in transplant medicine: a critical review of current approaches. Transplantation. 2016;100:2024–38. https://doi.org/10.1097/TP.0000000000001321.
Nesvizhskii AI, Vitek O, Aebersold R. Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat Methods. 2007;4(10):787–97. https://doi.org/10.1038/nmeth1088.
Tang N, Tornatore P, Weinberger SR. Current developments in SELDI affinity technology. Mass Spectrom Rev. 2004;23(1):34–44. https://doi.org/10.1002/mas.10066.
Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69(23):4751–60.
Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198. https://doi.org/10.1038/nature01511.
Sigdel TK, Kaushal A, Gritsenko M, Norbeck AD, Qian WJ, Xiao W, et al. Shotgun proteomics identifies proteins specific for acute renal transplant rejection. Proteom Clin Appl. 2010;4(1):32–47. https://doi.org/10.1002/prca.200900124.
Xie F, Liu T, Qian W-J, Petyuk VA, Smith RD. Liquid chromatography-mass spectrometry-based quantitative proteomics. J Biol Chem. 2011;286(29):25443–9. https://doi.org/10.1074/jbc.R110.199703.
Sigdel TK, Gao Y, He J, Wang A, Nicora CD, Fillmore TL, et al. Mining the human urine proteome for monitoring renal transplant injury. Kidney Int. 2016;89(6):1244–52. https://doi.org/10.1016/j.kint.2015.12.049.
Ling XB, Sigdel TK, Lau K, Ying L, Lau I, Schilling J, et al. Integrative urinary peptidomics in renal transplantation identifies biomarkers for acute rejection. J Am Soc Nephrol. 2010;21(4):646–53. https://doi.org/10.1681/ASN.2009080876.
Han X, Aslanian A, Yates JR. Mass spectrometry for proteomics. Curr Opin Chem Biol. 2008;12(5):483–90. https://doi.org/10.1016/j.cbpa.2008.07.024.
Karas M, Bachmann D, Bahr U, Hillenkamp F. Matrix-assisted ultraviolet laser desorption of non-volatile compounds. Int J Mass Spectrom Ion Process. 1987;78:53–68. https://doi.org/10.1016/0168-1176(87)87041-6.
Freue GV, Sasaki M, Meredith A, Gunther OP, Bergman A, Takhar M, et al. Proteomic signatures in plasma during early acute renal allograft rejection. Mol Cell Proteom. 2010;9(9):1954–67. https://doi.org/10.1074/mcp.M110.000554.
Wegdam W, Moerland PD, Meijer D, de Jong SM, Hoefsloot HCJ, Kenter GG, et al. A critical assessment of SELDI-TOF-MS for biomarker discovery in serum and tissue of patients with an ovarian mass. Proteome Sci. 2012;10:45. https://doi.org/10.1186/1477-5956-10-45.
Baggerly KA, Morris JS, Coombes KR. Reproducibility of SELDI-TOF protein patterns in serum: comparing datasets from different experiments. Bioinformatics. 2004;20(5):777–85. https://doi.org/10.1093/bioinformatics/btg484.
Metzger J, Chatzikyrkou C, Broecker V, Schiffer E, Jaensch L, Iphoefer A, et al. Diagnosis of subclinical and clinical acute T-cell-mediated rejection in renal transplant patients by urinary proteome analysis. Proteom Clin Appl. 2011;5(5–6):322–33. https://doi.org/10.1002/prca.201000153.
Huang H, Xu X, Yao C, Cai M, Qian Y, Wang X, et al. Serum levels of CXCR3 ligands predict T cell-mediated acute rejection after kidney transplantation. Mol Med Rep. 2014;9(1):45–50. https://doi.org/10.3892/mmr.2013.1753.
Hricik DE, Nickerson P, Formica RN, Poggio ED, Rush D, Newell KA, et al. Multicenter validation of urinary CXCL9 as a risk-stratifying biomarker for kidney transplant injury. Am J Transplant. 2013;13(10):2634–44. https://doi.org/10.1111/ajt.12426.
Hirt-Minkowski P, Amico P, Ho J, Gao A, Bestland J, Hopfer H, et al. Detection of clinical and subclinical tubulo-interstitial inflammation by the urinary CXCL10 chemokine in a real-life setting. Am J Transplant. 2012;12(7):1811–23. https://doi.org/10.1111/j.1600-6143.2012.03999.x.
Raza A, Firasat S, Khaliq S, Aziz T, Mubarak M, Naqvi SA, et al. The association of urinary interferon-gamma inducible protein-10 (IP10/CXCL10) levels with kidney allograft rejection. Inflamm Res. 2017;66(5):425–32. https://doi.org/10.1007/s00011-017-1025-7.
Ho J, Sharma A, Mandal R, Wishart DS, Wiebe C, Storsley L, et al. Detecting renal allograft inflammation using quantitative urine metabolomics and CXCL10. Transplant Direct. 2016;2(6):e78. https://doi.org/10.1097/txd.0000000000000589.
Crespo E, Cravedi P, Martorell J, Luque S, Melilli E, Cruzado JM, et al. Posttransplant peripheral blood donor-specific interferon-gamma enzyme-linked immune spot assay differentiates risk of subclinical rejection and de novo donor-specific alloantibodies in kidney transplant recipients. Kidney Int. 2017;92(1):201–13. https://doi.org/10.1016/j.kint.2016.12.024.
De Serres SA, Mfarrej BG, Grafals M, Riella LV, Magee CN, Yeung MY, et al. Derivation and validation of a cytokine-based assay to screen for acute rejection in renal transplant recipients. Clin J Am Soc Nephrol. 2012;7(6):1018–25. https://doi.org/10.2215/cjn.11051011.
Fabrega E, Lopez-Hoyos M, San Segundo D, Casafont F, Angel Mieses M, Sampedro B, et al. Serum levels of interleukin-9 during acute rejection in liver transplantation. Transplant Proc. 2012;44(6):1533–5. https://doi.org/10.1016/j.transproceed.2012.05.013.
Fabrega E, Lopez-Hoyos M, San Segundo D, Casafont F, Pons-Romero F. Changes in the serum levels of interleukin-17/interleukin-23 during acute rejection in liver transplantation. Liver Transplant. 2009;15(6):629–33. https://doi.org/10.1002/lt.21724.
Sharma VK, Bologa RM, Li B, Xu GP, Lagman M, Hiscock W, et al. Molecular executors of cell death–differential intrarenal expression of Fas ligand, Fas, granzyme B, and perforin during acute and/or chronic rejection of human renal allografts. Transplantation. 1996;62(12):1860–6.
Heng B, Ding H, Ren H, Shi L, Chen J, Wu X, et al. Diagnostic performance of Fas ligand mRNA expression for acute rejection after kidney transplantation: a systematic review and meta-analysis. PLoS One. 2016;11(11):e0165628-e. https://doi.org/10.1371/journal.pone.0165628.
Lettau M, Paulsen M, Schmidt H, Janssen O. Insights into the molecular regulation of FasL (CD178) biology. Eur J Cell Biol. 2011;90(6–7):456–66. https://doi.org/10.1016/j.ejcb.2010.10.006.
Vasconcellos LM, Schachter AD, Zheng XX, Vasconcellos LH, Shapiro M, Harmon WE, et al. Cytotoxic lymphocyte gene expression in peripheral blood leukocytes correlates with rejecting renal allografts. Transplantation. 1998;66(5):562–6.
Roedder S, Sigdel T, Salomonis N, Hsieh S, Dai H, Bestard O, et al. The kSORT assay to detect renal transplant patients at high risk for acute rejection: results of the multicenter AART study. PLoS Med. 2014;11(11):e1001759. https://doi.org/10.1371/journal.pmed.1001759.
Li L, Khatri P, Sigdel TK, Tran T, Ying L, Vitalone MJ, et al. A peripheral blood diagnostic test for acute rejection in renal transplantation. Am J Transplant. 2012;12(10):2710–8. https://doi.org/10.1111/j.1600-6143.2012.04253.x.
Crespo E, Roedder S, Sigdel T, Hsieh SC, Luque S, Cruzado JM, et al. Molecular and functional noninvasive immune monitoring in the ESCAPE study for prediction of subclinical renal allograft rejection. Transplantation. 2017;101(6):1400–9. https://doi.org/10.1097/tp.0000000000001287.
Li B, Hartono C, Ding R, Sharma VK, Ramaswamy R, Qian B, et al. Noninvasive diagnosis of renal-allograft rejection by measurement of messenger RNA for perforin and granzyme B in urine. N Engl J Med. 2001;344(13):947–54. https://doi.org/10.1056/NEJM200103293441301.
Suthanthiran M, Schwartz JE, Ding R, Abecassis M, Dadhania D, Samstein B, et al. Urinary-cell mRNA profile and acute cellular rejection in kidney allografts. N Engl J Med. 2013;369(1):20–31. https://doi.org/10.1056/NEJMoa1215555.
Meersch M, Schmidt C, Van Aken H, Martens S, Rossaint J, Singbartl K, et al. Urinary TIMP-2 and IGFBP7 as early biomarkers of acute kidney injury and renal recovery following cardiac surgery. PLoS One. 2014;9(3):e93460-e. https://doi.org/10.1371/journal.pone.0093460.
Galichon P, Amrouche L, Hertig A, Brocheriou I, Rabant M, Xu-Dubois YC, et al. Urinary mRNA for the diagnosis of renal allograft rejection: the issue of normalization. Am J Transplant. 2016;16(10):3033–40. https://doi.org/10.1111/ajt.13891.
Seo JW, Moon H, Kim SY, Moon JY, Jeong KH, Lee YH, et al. Both absolute and relative quantification of urinary mRNA are useful for non-invasive diagnosis of acute kidney allograft rejection. PLoS One. 2017;12(6):e0180045. https://doi.org/10.1371/journal.pone.0180045.
Muthukumar T, Dadhania D, Ding R, Snopkowski C, Naqvi R, Lee JB, et al. Messenger RNA for FOXP3 in the urine of renal-allograft recipients. N Engl J Med. 2005;353(22):2342–51. https://doi.org/10.1056/NEJMoa051907.
Afaneh C, Muthukumar T, Lubetzky M, Ding R, Snopkowski C, Sharma VK, et al. Urinary cell levels of mRNA for OX40, OX40L, PD-1, PD-L1, or PD-L2 and acute rejection of human renal allografts. Transplantation. 2010;90(12):1381–7. https://doi.org/10.1097/TP.0b013e3181ffbadd.
Medeiros M, Sharma VK, Ding R, Yamaji K, Li B, Muthukumar T, et al. Optimization of RNA yield, purity and mRNA copy number by treatment of urine cell pellets with RNAlater. J Immunol Methods. 2003;279(1–2):135–42.
Waikar SS, Betensky RA, Emerson SC, Bonventre JV. Imperfect gold standards for kidney injury biomarker evaluation. J Am Soc Nephrol. 2012;23(1):13–21. https://doi.org/10.1681/asn.2010111124.
Pickering JW, Endre ZH. Linking injury to outcome in acute kidney injury: a matter of sensitivity. PLoS One. 2013;8(4):e62691. https://doi.org/10.1371/journal.pone.0062691.
Lorenzen JM, Volkmann I, Fiedler J, Schmidt M, Scheffner I, Haller H, et al. Urinary miR-210 as a mediator of acute T-cell mediated rejection in renal allograft recipients. Am J Transplant. 2011;11(10):2221–7. https://doi.org/10.1111/j.1600-6143.2011.03679.x.
De Vlaminck I, Valantine HA, Snyder TM, Strehl C, Cohen G, Luikart H, et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med. 2014;6(241):241ra77. https://doi.org/10.1126/scitranslmed.3007803.
De Vlaminck I, Martin L, Kertesz M, Patel K, Kowarsky M, Strehl C, et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proc Natl Acad Sci USA. 2015;112(43):13336–41. https://doi.org/10.1073/pnas.1517494112.
Johnston O, Cassidy H, O’Connell S, O’Riordan A, Gallagher W, Maguire PB, et al. Identification of beta2-microglobulin as a urinary biomarker for chronic allograft nephropathy using proteomic methods. Proteom Clin Appl. 2011;5(7–8):422–31. https://doi.org/10.1002/prca.201000160.
Kurian SM, Heilman R, Mondala TS, Nakorchevsky A, Hewel JA, Campbell D, et al. Biomarkers for early and late stage chronic allograft nephropathy by proteogenomic profiling of peripheral blood. PLoS One. 2009;4(7):e6212. https://doi.org/10.1371/journal.pone.0006212.
Banon-Maneus E, Diekmann F, Carrascal M, Quintana LF, Moya-Rull D, Bescos M, et al. Two-dimensional difference gel electrophoresis urinary proteomic profile in the search of nonimmune chronic allograft dysfunction biomarkers. Transplantation. 2010;89(5):548–58. https://doi.org/10.1097/TP.0b013e3181c690e3.
Maluf DG, Dumur CI, Suh JL, Scian MJ, King AL, Cathro H, et al. The urine microRNA profile may help monitor post-transplant renal graft function. Kidney Int. 2014;85(2):439–49. https://doi.org/10.1038/ki.2013.338.
Soltaninejad E, Nicknam MH, Nafar M, Sharbafi MH, Keshavarz Shahbaz S, Barabadi M, et al. Altered expression of microRNAs following chronic allograft dysfunction with interstitial fibrosis and tubular atrophy. Iran J Allergy Asthma Immunol. 2015;14(6):615–23.
Iwasaki K, Yamamoto T, Inanaga Y, Hiramitsu T, Miwa Y, Murotani K, et al. MiR-142-5p and miR-486-5p as biomarkers for early detection of chronic antibody-mediated rejection in kidney transplantation. Biomarkers. 2017;22(1):45–54. https://doi.org/10.1080/1354750x.2016.1204000.
Mas V, Maluf D, Archer K, Yanek K, Mas L, King A, et al. Establishing the molecular pathways involved in chronic allograft nephropathy for testing new noninvasive diagnostic markers. Transplantation. 2007;83(4):448–57. https://doi.org/10.1097/01.tp.0000251373.17997.9a.
Hope CM, Troelnikov A, Hanf W, Jesudason S, Coates PT, Heeger PS, et al. Peripheral natural killer cell and allo-stimulated T-cell function in kidney transplant recipients associate with cancer risk and immunosuppression-related complications. Kidney Int. 2015;88(6):1374–82. https://doi.org/10.1038/ki.2015.237.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
ZHE has received travel support from Ortho Diagnostics, Roche Pharmaceuticals and the Novartis Australian Renal Transplant Advisory Board. SH, JE and AYMA declare that they have no conflicts of interest.
Funding
ZHE has received research funding from the Australian National Health and Medical Research Council and the New Zealand Health Research Council. SH has received funding from the Prince of Wales clinical school scholarship, Australia.
Rights and permissions
About this article
Cite this article
Herath, S., Erlich, J., Au, A.Y.M. et al. Advances in Detection of Kidney Transplant Injury. Mol Diagn Ther 23, 333–351 (2019). https://doi.org/10.1007/s40291-019-00396-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-019-00396-z