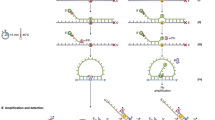
Abstract
BEAMing polymerase chain reaction (PCR) and digital PCR (dPCR) are used for robust and accurate quantification of nucleic acids. These methods are particularly well suited for the identification of very small fractions (<1%) of variant copies such as the presence of mutant genes in a predominantly wild-type background. BEAMing and dPCR are increasingly used in diverse fields including bacteriology, virology, non-invasive prenatal testing, and oncology, in particular for the molecular analysis of liquid biopsies. In this review, we present the principles of BEAMing and dPCR as well as the trends of future technical development, focusing on the possibility of developing multiplexed mutation analysis. Finally, we will discuss why such techniques will remain useful despite the ever-decreasing costs and increased automatization of next-generation sequencing (NGS), using molecular characterization of cancer cells as an example.



Similar content being viewed by others
References
Sykes PJ, Neoh SH, Brisco MJ, Hughes E, Condon J, Morley AA. Quantitation of targets for PCR by use of limiting dilution. Biotechniques. 1992;13(3):444–9.
Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci USA. 1999;96(16):9236–41.
Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci USA. 2003;100(15):8817–22.
Li M, Diehl F, Dressman D, Vogelstein B, Kinzler KW. BEAMing up for detection and quantification of rare sequence variants. Nat Methods. 2006;3(2):95–7.
Diehl F, Li M, He Y, Kinzler KW, Vogelstein B, Dressman D. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat Methods. 2006;3(7):551–9.
Benesova L, Belsanova B, Suchanek S, Kopeckova M, Minarikova P, Lipska L, et al. Mutation-based detection and monitoring of cell-free tumor DNA in peripheral blood of cancer patients. Anal Biochem. 2013;433(2):227–34.
Tabernero J, Lenz HJ, Siena S, Sobrero A, Falcone A, Ychou M, et al. Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: a retrospective, exploratory analysis of the CORRECT trial. Lancet Oncol. 2015;16(8):937–48.
Thress KS, Brant R, Carr TH, Dearden S, Jenkins S, Brown H, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: a cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer. 2015;90(3):509–15.
Sorber L, Zwaenepoel K, Deschoolmeester V, Van Schil PE, Van Meerbeeck J, Lardon F, et al. Circulating cell-free nucleic acids and platelets as a liquid biopsy in the provision of personalized therapy for lung cancer patients. Lung Cancer. 2017;107:100–107.
Vynck M, Trypsteen W, Thas O, Vandekerckhove L, De Spiegelaere W. The future of digital polymerase chain reaction in virology. Mol Diagn Ther. 2016;20(5):437–47.
Doi H, Uchii K, Takahara T, Matsuhashi S, Yamanaka H, Minamoto T. Use of droplet digital PCR for estimation of fish abundance and biomass in environmental DNA surveys. PLoS One. 2015;10(3):e0122763.
El Khattabi LA, Rouillac-Le Sciellour C, Le Tessier D, Luscan A, Coustier A, Porcher R, et al. Could digital PCR be an alternative as a non-invasive prenatal test for trisomy 21: a proof of concept study. PLoS One. 2016;11(5):e0155009.
Hudecova I. Digital PCR analysis of circulating nucleic acids. Clin Biochem. 2015;48(15):948–56.
Allenson K, Castillo J, San Lucas FA, Scelo G, Kim DU, Bernard V, et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol. 2017;28(4):741–7.
Denis JA, Patroni A, Guillerm E, Pepin D, Benali-Furet N, Wechsler J, et al. Droplet digital PCR of circulating tumor cells from colorectal cancer patients can predict KRAS mutations before surgery. Mol Oncol. 2016;10(8):122.
Janku F, Huang HJ, Fujii T, Shelton DN, Madwani K, Fu S, et al. Multiplex KRASG12/G13 mutation testing of unamplified cell-free DNA from the plasma of patients with advanced cancers using droplet digital polymerase chain reaction. Ann Oncol. 2016;28(3):642–50.
Pender A, Garcia-Murillas I, Rana S, Cutts RJ, Kelly G, Fenwick K, et al. Efficient genotyping of KRAS mutant non-small cell lung cancer using a multiplexed droplet digital PCR approach. PLoS One. 2015;10(9):e0139074.
Taly V, Pekin D, Benhaim L, Kotsopoulos SK, Le Corre D, Li X, et al. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin Chem. 2013;59(12):172.
Zonta E, Garlan F, Pecuchet N, Perez-Toralla K, Caen O, Milbury C, et al. Multiplex detection of rare mutations by picoliter droplet based digital PCR: sensitivity and specificity considerations. PLoS One. 2016;11(7):e015909.
Madic J, Zocevic A, Senlis V, Fradet E, Andre B, Muller S, et al. Three-color crystal digital PCR. Biomol Detect Quantif. 2016;10:34–46.
Wang M, Escudero-Ibarz L, Moody S, Zeng N, Clipson A, Huang Y, et al. Somatic mutation screening using archival formalin-fixed, paraffin-embedded tissues by Fluidigm multiplex PCR and Illumina sequencing. J Mol Diagn. 2015;17(5):521–32.
Whale AS, Huggett JF, Tzonev S. Fundamentals of multiplexing with digital PCR. Biomol Detect Quantif. 2016;10:15–23.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74.
Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–58.
Vogelstein B, Kinzler KW. The path to cancer—three strikes and you’re out. N Engl J Med. 2015;373(20):1895–8.
Tomasetti C, Marchionni L, Nowak MA, Parmigiani G, Vogelstein B. Only three driver gene mutations are required for the development of lung and colorectal cancers. Proc Natl Acad Sci USA. 2015;112(1):118–23.
Escargueil AE, Prado S, Dezaire A, Clairambault J, Larsen AK, Soares DG. Genotype- or phenotype-targeting anticancer therapies? Lessons from tumor evolutionary biology. Curr Pharm Des. 2016;22:6625–44.
Tafe LJ. Molecular mechanisms of therapy resistance in solid tumors: chasing “moving” targets. Virchows Arch. 2017. doi:10.1007/s00428-017-2101-7
Saad N, Poudel A, Basnet A, Gajra A. Epidermal growth factor receptor T790M mutation-positive metastatic non-small-cell lung cancer: focus on osimertinib (AZD9291). Onco Targets Ther. 2017;10:1757–66.
Araldi RP, Modolo DG, de Sa Junior PL, Consonni SR, de Carvalho RF, Roperto FP, et al. Genetics and metabolic deregulation following cancer initiation: a world to explore. Biomed Pharmacother. 2016;82:449–58.
Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66(8):3992–5.
Chi KR. The tumour trail left in blood. Nature. 2016;532(7598):269–71.
Cai X, Janku F, Zhan Q, Fan JB. Accessing genetic information with liquid biopsies. Trends Genet. 2016;31(10):564–75.
Speicher MR, Pantel K. Tumor signatures in the blood. Nat Biotechnol. 2014;32(5):441–3.
Cheng F, Su L, Qian C. Circulating tumor DNA: a promising biomarker in the liquid biopsy of cancer. Oncotarget. 2016;7(30):48832–41.
Parsons HA, Beaver JA, Park BH. Circulating plasma tumor DNA. Adv Exp Med Biol. 2016;882:259–76.
Francis G, Stein S. Circulating cell-free tumour DNA in the management of cancer. Int J Mol Sci. 2016;16(6):14122–42.
Ma M, Zhu H, Zhang C, Sun X, Gao X, Chen G. “Liquid biopsy”-ctDNA detection with great potential and challenges. Ann Transl Med. 2015;3(16):235.
Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem. 2015;61(1):112–23.
Alix-Panabieres C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem. 2013;59(1):110–8.
Plaks V, Koopman CD, Werb Z. Cancer. Circulating tumor cells. Science. 2013;341(6151):1186–8.
Tkach M, Thery C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164(6):1226–32.
Liu Y, Cao X. Organotropic metastasis: role of tumor exosomes. Cell Res. 2016;26(2):149–50.
Rak J. Cancer: organ-seeking vesicles. Nature. 2015;527(7578):312–4.
Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–35.
Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339(6122):959–61.
Nault JC, Zucman-Rossi J. TERT promoter mutations in primary liver tumors. Clin Res Hepatol Gastroenterol. 2016;40(1):9–14.
Zucman-Rossi J, Villanueva A, Nault JC, Llovet JM. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology. 2015;149(5):1226–39 e4.
Tavares C, Melo M, Cameselle-Teijeiro JM, Soares P, Sobrinho-Simoes M. Endocrine tumours: genetic predictors of thyroid cancer outcome. Eur J Endocrinol. 2016;174(4):R117–26.
Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–508.
Killela PJ, Pirozzi CJ, Healy P, Reitman ZJ, Lipp E, Rasheed BA, et al. Mutations in IDH1, IDH2, and in the TERT promoter define clinically distinct subgroups of adult malignant gliomas. Oncotarget. 2014;5(6):1515–25.
Huang A, Zhang X, Zhou SL, Cao Y, Huang XW, Fan J, et al. Detecting circulating tumor DNA in hepatocellular carcinoma patients using droplet digital PCR is feasible and reflects intratumoral heterogeneity. J Cancer. 2016;7(13):1907–14.
Luthra R, Patel KP, Routbort MJ, Broaddus RR, Yau J, Simien C, et al. A targeted high-throughput next-generation sequencing panel for clinical screening of mutations, gene amplifications, and fusions in solid tumors. J Mol Diagn. 2017;19(2):255–64.
Sheridan C. Milestone approval lifts Illumina’s NGS from research into clinic. Nat Biotechnol. 2014;32(2):111–2.
Kinugasa H, Nouso K, Tanaka T, Miyahara K, Morimoto Y, Dohi C, et al. Droplet digital PCR measurement of HER2 in patients with gastric cancer. Br J Cancer. 2015;112(10):1652–5.
Zhu Y, Lu D, Lira ME, Xu Q, Du Y, Xiong J, et al. Droplet digital polymerase chain reaction detection of HER2 amplification in formalin fixed paraffin embedded breast and gastric carcinoma samples. Exp Mol Pathol. 2016;100(2):287–93.
Wang Q, Yang X, He Y, Ma Q, Lin L, Fu P, et al. Droplet digital PCR for absolute quantification of EML4-ALK gene rearrangement in lung adenocarcinoma. J Mol Diagn. 2015;17(5):515–20.
Laurent-Puig P, Pekin D, Normand C, Kotsopoulos SK, Nizard P, Perez-Toralla K, et al. Clinical relevance of KRAS-mutated subclones detected with picodroplet digital PCR in advanced colorectal cancer treated with anti-EGFR therapy. Clin Cancer Res. 2015;21(5):1087–97.
Oxnard GR, Paweletz CP, Kuang Y, Mach SL, O’Connell A, Messineo MM, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res. 2014;20(6):1698–705.
Sanmamed MF, Fernandez-Landazuri S, Rodriguez C, Zarate R, Lozano MD, Zubiri L, et al. Quantitative cell-free circulating BRAFV600E mutation analysis by use of droplet digital PCR in the follow-up of patients with melanoma being treated with BRAF inhibitors. Clin Chem. 2015;61(1):297–304.
Watanabe M, Kawaguchi T, Isa S, Ando M, Tamiya A, Kubo A, et al. Ultra-sensitive detection of the pretreatment EGFR T790M mutation in non-small cell lung cancer patients with an EGFR-activating mutation using droplet digital PCR. Clin Cancer Res. 2015;21(15):3552–60.
Lee JY, Qing X, Xiumin W, Yali B, Chi S, Bak SH, et al. Longitudinal monitoring of EGFR mutations in plasma predicts outcomes of NSCLC patients treated with EGFR TKIs: Korean Lung Cancer Consortium (KLCC-12-02). Oncotarget. 2016;7(6):6984–93.
Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24.
Oellerich M, Schutz E, Beck J, Kanzow P, Plowman PN, Weiss GJ, et al. Using circulating cell-free DNA to monitor personalized cancer therapy. Crit Rev Clin Lab Sci. 2017;10:1–14.
Brychta N, Krahn T, von Ahsen O. Detection of KRAS mutations in circulating tumor DNA by digital PCR in early stages of pancreatic cancer. Clin Chem. 2016;62(11):1482–91.
Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci USA. 2005;102(45):16368–73.
Taly V, Pekin D, Benhaim L, Kotsopoulos SK, Le Corre D, Li X, et al. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin Chem. 2013;59(12):1722–31.
Normanno N, Denis MG, Thress KS, Ratcliffe M, Reck M. Guide to detecting epidermal growth factor receptor (EGFR) mutations in ctDNA of patients with advanced non-small-cell lung cancer. Oncotarget. 2017;8(7):12501–16.
Li Z, Zhang Y, Bao W, Jiang C. Insufficiency of peripheral blood as a substitute tissue for detecting EGFR mutations in lung cancer: a meta-analysis. Target Oncol. 2014;9(4):381–8.
Zheng D, Ye X, Zhang MZ, Sun Y, Wang JY, Ni J, et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci Rep. 2016;12(6):20913.
Alix-Panabieres C, Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6(5):479–91.
Lohr JG, Adalsteinsson VA, Cibulskis K, Choudhury AD, Rosenberg M, Cruz-Gordillo P, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol. 2014;32(5):479–84.
Shaw JA, Guttery DS, Hills A, Fernandez-Garcia D, Page K, Rosales BM, et al. Mutation analysis of cell-free DNA and single circulating tumor cells in metastatic breast cancer patients with high CTC counts. Clin Cancer Res. 2016;23(1):88–96.
Pestrin M, Salvianti F, Galardi F, De Luca F, Turner N, Malorni L, et al. Heterogeneity of PIK3CA mutational status at the single cell level in circulating tumor cells from metastatic breast cancer patients. Mol Oncol. 2015;9(4):749–57.
Fernandez SV, Bingham C, Fittipaldi P, Austin L, Palazzo J, Palmer G, et al. TP53 mutations detected in circulating tumor cells present in the blood of metastatic triple negative breast cancer patients. Breast Cancer Res. 2014;16(5):445.
Gasch C, Pantel K, Riethdorf S. Whole genome amplification in genomic analysis of single circulating tumor cells. Methods Mol Biol. 2015;1347:221–32.
De Luca F, Rotunno G, Salvianti F, Galardi F, Pestrin M, Gabellini S, et al. Mutational analysis of single circulating tumor cells by next generation sequencing in metastatic breast cancer. Oncotarget. 2016;7(18):26107–19.
Huggett JF, Cowen S, Foy CA. Considerations for digital PCR as an accurate molecular diagnostic tool. Clin Chem. 2015;61(1):79–88.
Huggett JF, Foy CA, Benes V, Emslie K, Garson JA, Haynes R, et al. The digital MIQE guidelines: minimum information for publication of quantitative digital PCR experiments. Clin Chem. 2013;59(6):892–902.
Stahlberg A, Krzyzanowski PM, Jackson JB, Egyud M, Stein L, Godfrey TE. Simple, multiplexed, PCR-based barcoding of DNA enables sensitive mutation detection in liquid biopsies using sequencing. Nucleic Acids Res. 2016;44(11):e105.
Albayrak C, Jordi CA, Zechner C, Lin J, Bichsel CA, Khammash M, et al. Digital quantification of proteins and mRNA in single mammalian cells. Mol Cell. 2016;61(6):914–24.
Sartore-Bianchi A, Pietrantonio F, Amatu A, Milione M, Cassingena A, Ghezzi S, et al. Digital PCR assessment of MGMT promoter methylation coupled with reduced protein expression optimises prediction of response to alkylating agents in metastatic colorectal cancer patients. Eur J Cancer. 2016;16(71):43–50.
Barault L, Amatu A, Bleeker FE, Moutinho C, Falcomata C, Fiano V, et al. Digital PCR quantification of MGMT methylation refines prediction of clinical benefit from alkylating agents in glioblastoma and metastatic colorectal cancer. Ann Oncol. 2015;26(9):1994–9.
Yu M, Carter KT, Makar KW, Vickers K, Ulrich CM, Schoen RE, et al. MethyLight droplet digital PCR for detection and absolute quantification of infrequently methylated alleles. Epigenetics. 2015;10(9):803–9.
Tian H, Sun Y, Liu C, Duan X, Tang W, Li Z. Precise quantitation of microRNA in a single cell with droplet digital PCR based on ligation reaction. Anal Chem. 2016;88(23):11384–11389.
Bellingham SA, Shambrook M, Hill AF. Quantitative analysis of exosomal miRNA via qPCR and digital PCR. Methods Mol Biol. 2017;1545:55–70.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
No specific funding was received for this article.
Conflict of interest
The authors (JAD, EG, FC, AKL & JML) declare no conflicts of interest.
Rights and permissions
About this article
Cite this article
Denis, J.A., Guillerm, E., Coulet, F. et al. The Role of BEAMing and Digital PCR for Multiplexed Analysis in Molecular Oncology in the Era of Next-Generation Sequencing. Mol Diagn Ther 21, 587–600 (2017). https://doi.org/10.1007/s40291-017-0287-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-017-0287-7