Abstract
Introduction
Vortioxetine is an antidepressant primarily metabolized by the polymorphic enzyme cytochrome P450 (CYP) 2D6. A population pharmacokinetic (popPK) model of vortioxetine and its CYP2D6-dependent metabolite was recently published.
Objective
The aim of the current study was to assess the predictive performance of the popPK model using vortioxetine concentration measurements from a clinical setting. Furthermore, the study aimed to evaluate the ability of different CYP2D6 phenotype classification systems to provide accurate concentration predictions.
Methods
Overall, 1388 patients receiving vortioxetine treatment were identified from a therapeutic drug monitoring (TDM) database in Oslo, Norway; 334 CYP2D6-genotyped patients with 502 serum concentrations of vortioxetine, analysed by a validated ultra-high-performance liquid chromatography–high-resolution mass spectrometry (UHPLC-HRMS) method, were retrospectively included. The performance of the vortioxetine popPK model was tested on the clinical data from the TDM database.
Results
Overall, the model had a good ability to predict vortioxetine concentrations measured in clinical practice, with a slight tendency to overpredict concentrations. Using simulation-based diagnostics, 76% of the prediction-corrected TDM concentrations were within the 90% prediction interval based on 1000 simulated data sets. Prediction-based diagnostics showed the best performance for CYP2D6 poor and ultrarapid metabolizers, with a median prediction error (MDPE) of 12% and 23%, respectively, while the poorest performance was observed for normal metabolizers, with an MDPE of 66%. In the comparison of different CYP2D6 phenotype classification systems, the use of differentiated activity scores for decreased function alleles did not improve the concentration predictions. Grouping the CYP2D6 genotypes into the four conventional phenotype groups provided predictions closest to the TDM measured concentrations.
Conclusion
TDM data provide a unique insight into real-world clinical practice with vortioxetine. The tendency of the popPK model to overpredict vortioxetine concentrations measured in TDM may be attributed to several factors, including poor treatment compliance for some patients and, to a lesser extent, lack of information on patient characteristics and misspecified CYP2D6 alleles. To optimize personalized therapy with vortioxetine, real-world clinical data sets originating from different ethnicities need to be studied in the future.
Similar content being viewed by others
The previously published population pharmacokinetic (popPK) model showed a good ability to predict vortioxetine concentrations measured in a clinical setting. |
The conventional cytochrome P450 (CYP) 2D6 phenotype classification system (poor metabolizers, intermediate metabolizers, normal metabolizers, ultrarapid metabolizers) was adequate for predicting vortioxetine exposure. |
The use of differentiated activity scores for decreased function alleles did not improve concentration predictions for vortioxetine. |
1 Introduction
All therapeutic drugs exhibit pharmacokinetic (PK) variability, and an understanding of this variability is fundamental to practising personalized medicine. Population PK (popPK) modelling can be used to quantify PK variability between individuals and to estimate the doses required for individual patients to obtain target drug concentrations [1].
Individual variability in phenotypes of drug-metabolizing enzymes is considered a key factor for interpatient PK variability. The cytochrome P450 (CYP) 2D6 enzyme is highly polymorphic, which causes substantial variability in enzyme activity between individuals [2]. More than 100 CYP2D6 allele variants have been identified to date, and, together with gene duplications, these give rise to (1) no, (2) decreased, (3) normal, or (4) increased enzyme activity [3].
To guide dose selection of CYP2D6-metabolized drugs, patients can be assigned a CYP2D6 phenotype based on their genotype. Several CYP2D6 genotype–phenotype classification systems exist, but most commonly patients are allocated to four different genotype-predicted phenotype subgroups: poor metabolizers (PMs), intermediate metabolizers (IMs), normal metabolizers (NMs) and ultrarapid metabolizers (UMs). However, critics find this system too simple and consequently a CYP2D6 activity score system was developed 10 years ago to improve the precision of predicting CYP2D6 metabolizer phenotype based on genotype [4, 5]. This system assigns a numeric value between 0 and 1 to each CYP2D6 allele reflecting the enzyme activity relative to the wild-type allele (CYP2D6*1). In the case of gene duplication, the score assigned to an allele is doubled. The activity score of a given CYP2D6 diplotype/genotype is then calculated as the sum of the values assigned to each allele [4]. However, this system has been a matter of debate, especially regarding the activity scores assigned to the decreased function alleles (e.g. CYP2D6*9, CYP2D6*10, CYP2D6*41).
Numerous antidepressants are metabolized by CYP2D6, and the large interindividual variability in enzyme activity results in considerable differences in drug exposure between patients [6]. A good understanding of the effect of CYP2D6 polymorphisms on drug PK is thus essential to optimize antidepressant drug therapy for the individual patient.
Vortioxetine is an antidepressant with a multimodal mechanism of action, indicated for the treatment of major depressive disorder (MDD) at oral doses of 5–20 mg/day. In clinical studies, vortioxetine has demonstrated a dose–response relationship, with higher doses yielding better efficacy on depressive symptoms [7], while nausea was the only adverse event showing dose-dependency [8]. In clinical practice, therapeutic drug monitoring (TDM) can be used as a tool to ensure vortioxetine exposure is maintained within a therapeutic reference window of approximately 10–40 ng/mL [9], thereby facilitating safe and efficacious pharmacotherapy.
Vortioxetine is primarily metabolized via CYP2D6 and, to a lesser extent, by the polymorphic enzyme CYP2C19 [10, 11]. In a recent study by Frederiksen et al., a popPK model was developed that quantified the CYP2D6-mediated metabolism of vortioxetine in more than 1100 healthy subjects carrying different CYP2D6 genotypes [12]. The model was developed based on data originating from phase I studies, i.e. a controlled clinical trial setting.
In order to assess the validity of the recent popPK model in a real-world setting, the current study aimed to evaluate the predictive performance of the popPK model using patient data from a TDM service. Furthermore, the study aimed to assess the ability of different CYP2D6 phenotype classification systems to accurately predict vortioxetine concentrations measured in clinical practice.
2 Methods
2.1 Study Population
Patients were retrospectively included in the study from the TDM database (Swisslab II, Roche Diagnostics, Berlin, Germany) at the Center for Psychopharmacology, Diakonhjemmet Hospital, Oslo, Norway. In Norway, TDM is commonly used within psychiatry as a tool for tailoring drug treatment and clinical follow-up. The laboratory service at the Center for Psychopharmacology annually performs a total of 40,000 TDM analyses of various psychiatric drugs, as well as CYP genotyping of approximately 5000 patients. The analyses are requested both in primary health care and in hospital settings. Outpatient requests comprise around 75% of all analyses. The data material is therefore representative for the country’s psychiatric health care practice.
In the present study, a search in the TDM database was performed to identify patients treated with vortioxetine during the period January 2013 to June 2020. Patients were included in the study if (1) they had been genotyped for CYP2D6 and (2) if they had a blood serum concentration of vortioxetine measured as part of routine clinical practice.
Information on prescribed vortioxetine dose, concomitant medication, and the time interval between the last vortioxetine dose and the TDM blood sample was retrieved from the TDM requisition forms. Patients were excluded if (1) vortioxetine was not detected in blood serum (reflecting complete noncompliance), or (2) information on vortioxetine dose or time between the last dose and blood sampling was missing. Furthermore, patients co-treated with CYP2D6 inhibitors (bupropion, fluoxetine, paroxetine and levomepromazine) or multiple-enzyme inducers (carbamazepine, phenobarbital, phenytoin, fluvoxamine), as determined by TDM measurements or information on the TDM requisition forms, were excluded.
The study was approved by the Regional Committee for Medical and Health Research Ethics and the Hospital Investigational Review Board.
2.2 Genotyping
CYP2D6 and CYP2C19 genotyping was performed using Taqman-based real-time polymerase chain reaction (PCR) assays at the Center for Psychopharmacology, Diakonhjemmet Hospital, Oslo, Norway.
The following CYP2D6 variant alleles were included in the genotyping panel: CYP2D6*3 (rs35742686), CYP2D6*4 (rs3892097), CYP2D6*5 (whole gene deletion), CYP2D6*6 (rs5030655), CYP2D6*9 (rs5030656), CYP2D6*10 (rs1065852), CYP2D6*41 (rs28371725), as well as copy number variation. These are standard variants included in clinical CYP2D6 genotyping panels, and, for example, predict > 90% of Caucasian PMs [13, 14]. Wild-type (CYP2D6*1) was assigned when no variant alleles were detected. Patients who carried genotypes with gene duplications combined with the detection of null or reduced function alleles, e.g. CYP2D6*1/4XN, were excluded from the analysis as the genotyping assay did not identify which allele was duplicated.
The CYP2C19 genotyping assay included the non-functional alleles CYP2C19*2 (rs4244285), CYP2C19*3 (rs4986893/rs57081121), CYP2C19*4 (rs28399504), and the increased function allele CYP2C19*17 (rs12248560).
CYP2D6 and CYP2C19 genotypes were translated into phenotypes according to recent consensus recommendations from the Clinical Pharmacogenetics Implementation Consortium (CPIC) and the Dutch Pharmacogenetics Working Group (DPWG) [15, 16].
2.3 Serum Concentration Assay
The serum concentration of vortioxetine was determined using an ultra-high-performance liquid chromatography–high-resolution mass spectrometry (UHPLC-HRMS) method validated and certified for routine TDM analyses at the Center for Psychopharmacology. The serum samples were purified by protein precipitation mixing 200 µL of serum aliquot with 400 µL of acetonitrile–methanol (90/10 vol/vol), which included the internal standard (isotope-labelled 13C6-vortioxetine). Following centrifugation for 10 min (4000 rpm at 4°C), 4 µL of purified sample was injected into the liquid chromatography tandem mass spectrometry (LC-MS/MS) system for vortioxetine analysis. The LC system was a Vanquish-UHPLC (Thermo Fisher Scientific, Waltham, MA, USA), and chromatographic separation was performed using an XBridge BEH C18-column (2.6 µm, 2.1 × 75 mm; Waters Corporation, Milford, MA, USA) kept at 35 °C. The mobile phase consisted of a gradient mixture of ammonium acetate (pH 4.8) and acetonitrile, and the flow rate was 0.6 mL/min, providing a retention time of 2.44 min for vortioxetine and 13C6-vortioxetine (total run time 4 min). Detection used a Q Exactive Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific) operating in positive ionization mode acquiring full scan data at a resolution of 70,000 (full width at half maximum at 200 Da). Simultaneously, data-dependent MS/MS (ddMS2) spectra was acquired (resolution of 17,500) for those precursor ions in the inclusion list. The full scan data were used for quantification, while the ddMS2 spectra allowed the confirmation of detected vortioxetine in the samples.
The lower limit of quantification (LLOQ) and the upper limit of quantification (ULOQ) of the assay were 5 nM and 195 nM, respectively. The intra- and interday inaccuracy was ≤ 12% at LLOQ and ≤ 3% at ULOQ, while the intra- and interday imprecision was ≤ 8% at LLOQ and ≤ 3% at ULOQ.
2.4 Population Pharmacokinetic Model Validation
PopPK modelling and simulations were performed in NONMEM (version 7.4; ICON Development Solutions, Ellicott City, MD, USA). The NONMEM output was processed using R software (version 3.5.1) run under R Studio (The R Foundation for Statistical Computing, Vienna, Austria).
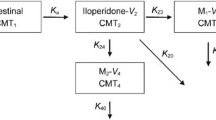
A popPK model describing the joint PK of vortioxetine and its CYP2D6-dependent metabolite was rebuilt as reported by Frederiksen et al. [12]. Briefly, the model was a six-compartment model including central and peripheral compartments for both vortioxetine and the metabolite, and a hypothetical absorption compartment to account for presystemic metabolite formation (quantified by the parameter Fmet). The total clearance of vortioxetine was divided into a non-CYP2D6-mediated (CLother) and a CYP2D6-mediated (CLCYP2D6) clearance, the latter reflecting the systemic formation of the metabolite. The extent of metabolite formation, as quantified by Fmet and CLCYP2D6, was used as a measure of CYP2D6 activity in the individual patient. It should be noted that formation of the metabolite could be driven by other enzymes/processes in addition to CYP2D6, and therefore the individual values of Fmet and CLCYP2D6 estimated for CYP2D6 PMs were >0, although these patients were expected to exhibit no CYP2D6 activity.
The ability of the popPK model to predict vortioxetine concentrations measured as part of routine clinical TDM was assessed by simulation- and prediction-based diagnostics.
2.4.1 Simulation-Based Diagnostics
Using the population parameter estimates (fixed and random effects) reported in the original popPK study [12], 1000 simulated datasets were generated, each corresponding to the data obtained from the TDM database. As the TDM data originated from several dose levels, prediction-corrected concentrations were calculated for the TDM observations and simulated model predictions using the following formula (Eq. 1):
where \({Y}_{ij}\) is the concentration for the ith individual at the jth time point, \({pcY}_{ij}\) is the prediction-corrected concentration, \({\widetilde{PRED }}_{bin}\) is the median of the typical population prediction for each specific bin, and \({PRED}_{ij}\) is the typical population prediction for the ith individual at the jth time point.
A prediction-corrected visual predictive check (pcVPC) plot was created to compare the TDM concentrations with the simulated model predictions. The amount of TDM concentrations outside the 90% prediction interval (PI) was used to assess whether the popPK model was representative of the TDM data.
Furthermore, normalized prediction distribution errors (NPDE) were computed and assessed graphically.
2.4.2 Prediction-Based Diagnostics
The popPK model was used to predict concentrations for all TDM patients at sampling times identical to those of the TDM measurements. The individual concentration predictions took into account available patient covariates and CYP2D6 genotype.
Patients’ CYP2D6 genotype was incorporated into the model predictions by adjusting the two model parameters reflecting CYP2D6-mediated metabolism: Fmet and CLCYP2D6. The magnitudes of Fmet and CLCYP2D6 were adjusted for each patient by the CYP2D6 activity score assigned based on patients’ CYP2D6 genotypes according to the recent consensus recommendation from the CPIC/Dutch Pharmacogenetics Working Group (DPWG) [15] (see the CYP2D6 phenotype classification system III in Table 1). For details on how Fmet and CLCYP2D6 were adjusted for each genotype, please refer to the electronic supplementary material (ESM). All other model parameters were fixed to the population estimates as reported in the original popPK analysis [12].
The original popPK model included the following covariate relationships: CYP2C19 phenotype, age and lean body mass (LBM) on CLother, creatinine clearance on metabolite clearance (CLmet), and height and weight on volumes of distribution [12]. In the current study, information on patient age, sex and genotype-based CYP2C19 phenotype was retrieved from the TDM database. In line with the published popPK model, the effect of CYP2C19 phenotype and age on CLother was included in the model predictions. As information on patient height, body weight and LBM were not available from the TDM database, these covariates were imputed based on patient sex and were included in the model. Based on median values from the original popPK dataset, females were imputed with a height of 162 cm, body weight of 62 kg and LBM of 45 kg, while males were imputed with a height of 175 cm, body weight of 76 kg and LBM of 59 kg. Information on patients’ renal function was not available from the TDM database and therefore the median creatinine clearance (118 mL/min) from the original popPK data set was imputed for all patients.
To assess the predictive performance of the popPK model, a prediction error (PE) was calculated for each TDM measurement using the following equation (Eq. 2):
where Cpredi represents the ith individual predicted concentration and Cobsi represents the ith observed concentration from the TDM database. The median prediction error (MDPE) and median absolute prediction error (MAPE) were used to assess the precision and bias of the model predictions. The predictive performance was considered acceptable when MDPE ≤ ± 20% and MAPE ≤ 30%, as suggested by Glass et al. [17].
2.5 Evaluation of Different Cytochrome P450 (CYP) 2D6 Phenotype Classification Systems
Four different CYP2D6 phenotype classification systems were tested for their ability to provide accurate concentration predictions when incorporated into the popPK model. The objective of this comparison was to evaluate whether the activity score systems provided more accurate concentration predictions compared with the conventional phenotype groups (PM/IM/NM/UM), and whether differentiating the activity scores for decreased function alleles improved the individual concentration predictions. An overview of the activity scores assigned to CYP2D6 genotypes according to the four classifications systems is presented in Table 1.
The four CYP2D6 phenotype classification systems tested were:
-
I.
Conventional CYP2D6 phenotype groups (PM/IM/NM/UM) [assigned according to the consensus recommendation reported by Caudle et al. [15]]
-
II.
Activity score system A: all decreased function alleles assigned an equal value of 0.5 (equivalent to the previous CPIC definition as described by Caudle et al. [15])
-
III.
Activity score system B: decreased function alleles assigned a value of either 0.25 (CYP2D6*10) or 0.5 (all other alleles) [equivalent to the new consensus definition reported by Caudle et al. [15]]
-
IV.
Activity score system C: all decreased function alleles assigned different values (0.21–0.37) corresponding to the relative CYP2D6 activity estimated for each allele in the metabolism of vortioxetine (as reported by Frederiksen et al. [12])
Each system was incorporated into the popPK model by adjusting Fmet and CLCYP2D6 according to the activity score assigned to each CYP2D6 genotype (see the ESM for details). The four versions of the popPK model were then used to calculate individual concentration predictions at sampling times corresponding to the TDM measurements.
The ability of the four systems to provide accurate concentration predictions was assessed by the PE, MDPE and MAPE, as described above.
3 Results
3.1 Study Population
In total, 1388 patients receiving vortioxetine treatment were identified in the TDM database, of whom 516 patients had been CYP2D6 genotyped; 334 patients representing 502 vortioxetine concentration measurements were found eligible for inclusion in the study. Figure 1 shows a flowchart of patient inclusion and Table 2 presents a summary of the characteristics of the included patients and their respective TDM measurements.
The distribution of CYP2D6 phenotypes in the study population was similar to what is normally observed in a Caucasian population [18] and thus there did not appear to be any selection bias with regard to CYP2D6.
Overall, 268 of the TDM samples (53%) were within the therapeutic reference range reported for vortioxetine (10–40 ng/mL), while 191 and 43 samples were below and above the range, respectively. CYP2D6 NMs comprised 75% of the samples below the target range and 16% of the samples above. CYP2D6 IMs and PMs together comprised 24% of the samples below the target range and 84% of the samples above the target range. CYP2D6 UMs represented 2% of the samples below the target range and none of the samples above.
3.2 Simulation-Based Model Diagnostics
Of the 502 vortioxetine serum concentrations from the TDM database, 40 individual measurements (corresponding to 8%) were above the simulated 95th percentile and 81 measurements (corresponding to 16%) were below the simulated 5th percentile (see Fig. 2). The majority of the prediction-corrected TDM concentrations above the simulated 95th percentile originated from CYP2D6 PMs and IMs, while the prediction-corrected TDM concentrations below the simulated 5th percentile were primarily from CYP2D6 NMs. Although the proportion of measurements outside the 90% PI was higher than the expected 10%, the simulations from the popPK model generally captured the TDM concentrations well and many of the outliers were captured by the 95% confidence intervals around the simulated percentiles.
Prediction-corrected visual predictive check plot comparing observed vortioxetine concentrations from the TDM database with simulated concentrations based on the population pharmacokinetic model. The dashed lines represent the 5th, 50th and 95th percentiles of the prediction-corrected simulated concentrations and the shaded fields represent 95% confidence intervals around the percentiles. The circles represent the prediction-corrected TDM concentrations coloured by patient-predicted CYP2D6 phenotype. UM ultra-rapid metabolizer, NM normal metabolizer, IM intermediate metabolizer, PM poor metabolizer, TDM therapeutic drug monitoring, CYP cytochrome P450
The NPDEs showed a distribution within an acceptable deviation for a normal distribution and without any significant trend over time or predicted concentration (see electronic supplementary Fig. S1).
3.3 Prediction-Based Model Diagnostics
Figure 3 shows the vortioxetine concentrations from the TDM database (Cobs) plotted against the individual concentration predictions generated by the popPK model (Cpred) incorporating the effect of patients’ CYP2D6 genotypes and available covariates.
Individual model-predicted vortioxetine concentrations versus observed concentrations from therapeutic drug monitoring database. The colour of the circles indicates the patient CYP2D6 genotype. The black line represents the unity line indicating perfect agreement between the model predictions and observed concentrations, and the red line shows the trend of the data. CYP cytochrome P450, TDM therapeutic drug monitoring
There was generally good agreement between the model predictions and the TDM concentrations, although the model showed a tendency to overpredict the vortioxetine concentration. This tendency is also evident from Fig. 4, where a positive PE is observed for the majority of the concentration measurements.
Prediction error of the model-predicted vortioxetine concentrations by CYP2D6 phenotype classification group. The circles represent the prediction error of each concentration measurement and the colour of the circles indicates the patient CYP2D6 genotype. The boxes represent interquartile ranges and the lines within the boxes indicate the median prediction error. The dashed vertical line highlights 0% prediction error indicating perfect agreement between the model prediction and observed concentration. UM ultrarapid metabolizer, NM normal metabolizer, IM intermediate metabolizer, PM poor metabolizer, CYP cytochrome P450
The model showed the best predictive performance for CYP2D6 PMs and UMs, with MDPEs of 12% and 23%, respectively, and MAPEs of 46% and 23%, respectively. The CYP2D6 IMs had an MDPE of 38% and an MAPE of 45%, while the poorest predictive performance was seen for NMs, with an MDPE of 66% and an MAPE of 68%. The extreme outliers (defined as PE > 400%) were either carriers of CYP2D6*1/*1 (n = 8), CYP2D6*1/*null (n = 4) or CYP2D6*null/*41 (n = 1).
Figure 5 shows the individual PEs against (a) the observed TDM concentrations and (b) the time interval between the last dose and the TDM sample. There was a clear tendency of large PEs being associated with TDM concentrations below 10 ng/mL, while the magnitude of the PE did not appear to be related to the timing of the TDM sample.
Prediction error of the model-predicted vortioxetine concentrations against a observed therapeutic drug monitoring concentration of vortioxetine and b the time interval between the last vortioxetine dose and the therapeutic drug monitoring sample. The horizontal dashed lines highlight 0% prediction error indicating perfect agreement between the model prediction and observed concentration. UM ultrarapid metabolizer, NM normal metabolizer, IM intermediate metabolizer, PM poor metabolizer, CYP cytochrome P450
3.4 Evaluation of Different CYP2D6 Phenotype Classification Systems
The overall MDPEs for the four classification systems were I: 46%; II: 52%; III: 52%; and IV: 56%, while the MAPEs were I: 54%; II: 59%; III: 59%; and IV: 61%. The conventional phenotype groups (PM/IM/NM/UM, classification system I) thus showed the best predictive ability overall, although none of the four systems met the predefined criteria of MDPE ≤ ± 20% and MAPE ≤ 30%.
The PE associated with each of the four CYP2D6 phenotype classification systems is presented for CYP2D6 genotypes involving decreased function alleles in Fig. 6.
Prediction error of the model-predicted vortioxetine concentrations for four different CYP2D6 phenotype classification systems for CYP2D6 genotypes involving decreased function alleles. Boxes represent interquartile ranges and lines within the boxes indicate the median prediction error. The dashed line highlights 0% prediction error, indicating perfect agreement between the model prediction and observed concentration. CYP cytochrome P450
Only minor differences were observed between the four classification systems and there was no consistent trend in any of the systems showing a superior ability to accurately predict the vortioxetine concentrations measured by TDM. In general, the conventional phenotype groups (system I) provided vortioxetine concentration predictions closest to the TDM measured concentrations for many of the CYP2D6 genotypes involving decreased function alleles, while the systems using differentiated activity scores for decreased function alleles (systems III and IV) seemed to have the poorest predictive ability overall.
It should be noted that some of the CYP2D6 genotypes were only represented by a small number of individual carriers and therefore statistical comparison of the predictive ability of the classification systems for the individual genotypes was not possible based on the data.
4 Discussion
In this study, we set out to evaluate the predictive performance of a popPK model of vortioxetine and its metabolite using naturalistic patient data from TDM.
Overall, we found that the model had a good ability to predict vortioxetine concentrations measured as part of routine clinical TDM. This supports the applicability of the popPK model to predict vortioxetine exposures in a real-world clinical setting and underlines the validity of the model-based findings previously reported [12].
In the simulation-based diagnostics, the largest prediction-corrected concentrations were observed among CYP2D6 PMs and IMs, which is not surprising as these are known to have a reduced clearance of vortioxetine compared with CYP2D6 NMs [10]. To ensure target drug concentrations are obtained and maintained, routine TDM may be particularly beneficial for CYP2D6 PMs and IMs treated with vortioxetine. Furthermore, to avoid supratherapeutic concentrations of vortioxetine, it is recommended that CYP2D6 PMs are treated with a maximum dose of 10 mg/day, as described in the drug label [19].
The model showed good predictive performance with a slight tendency to overpredict vortioxetine concentrations measured by TDM.
The very large overpredictions were primarily observed among the low TDM concentration measurements across all CYP2D6 phenotype groups. A plausible explanation for these outliers is poor treatment compliance, which causes lower drug concentrations than expected based on the administered dose, and thereby high PEs.
The popPK model of vortioxetine and its metabolite was developed based on phase I data, where close to 100% treatment compliance may be assumed. In a real-world clinical setting, lower treatment compliance is common and particularly depression has been shown to be a risk factor for noncompliance [20]. In a study comparing PK data from vortioxetine phase I and phase II/III studies, a significant difference in vortioxetine exposure between the two study populations was observed [21]. As all patient factors known to affect variability in PK were corrected for in the study, the difference in vortioxetine exposure was thought to be attributed to noncompliance among the phase II/III study population (MDD patients). Compliance among the patients in the phase II/III studies was, on average, 75%, which corresponds well with the difference observed in exposure between the TDM data and model predictions in the current study. This may explain the higher PEs at lower versus higher vortioxetine concentrations.
Regarding the analytical method for determination of vortioxetine serum concentration, the validation parameters were excellent over the whole concentration range. Therefore, the analytical performance is unlikely to be relevant for the increased PEs at lower vortioxetine serum concentrations.
The discrepancy between the model predictions and observed TDM concentrations may also be explained by some of the limitations related to the nature of the TDM data. For example, the time intervals between the last dose and TDM sampling were based on patient reports and were therefore associated with some uncertainty. However, the data showed no systematic bias of the PEs over time, and, furthermore, as vortioxetine has a long elimination half-life, the effect of inaccuracies in the sampling time would only cause minor deviations in the model-predicted concentration.
Another limitation of the TDM data was the lack of information on some patient characteristics, such as body size measurements, and renal function. The effect of patients’ CYP2C19-predicted phenotype and age (as identified from the TDM database) on the model parameter CLother was used to calculate individual model predictions in the current study. However, patients’ body size measurements and creatinine clearance were imputed and therefore the effect of these covariates on the PK were not optimally captured in the model predictions, which could be a possible explanation for some of the discrepancy in the TDM measurements. However, the magnitude of the covariate effects in the popPK model was only small and should not affect the PK to a clinically significant extent. Therefore, the missing information on patient characteristics was unlikely to be the main explanation for the prediction error.
Furthermore, when the physicians/psychiatrists list co-prescribed drugs on the TDM requisition forms, drugs used for the treatment of somatic diseases may be missed. Thus, the ability to detect potential interactions with somatic drugs affecting vortioxetine serum concentration was lower than for CNS medications, which may also explain some of the prediction error.
Another potential issue is that the standard CYP2D6 genotyping panel at the laboratory service, developed for clinical use, only comprises the most common and relevant allele variants in Caucasians. These predict >90% of CYP2D6 PM [13, 14], but rare CYP2D6 variants may in some patients have contributed to mismatches between observed and predicted serum concentrations. More likely however is that the current knowledge on variants encoding the UM phenotype is limited, hence implying a possible reason for overpredicting the vortioxetine serum concentration among patients defined as CYP2D6*1/*1 carriers. This is supported by the large variability in vortioxetine concentrations and extreme PE outliers observed among CYP2D6*1/*1 patients, who are most likely to carry rare or unknown variants.
In the evaluation of different CYP2D6 phenotype classification systems, only minor differences were observed in the ability of the systems to predict TDM measured concentrations. The use of differentiated activity scores for decreased function alleles did not improve concentration predictions, although it should be noted that data on individual genotypes were limited.
Based on the results, the conventional phenotype classification system (PM/IM/NM/UM) appears to be adequate for the purpose of providing dose recommendations to obtain target concentrations of vortioxetine. The use of a more restricted number of phenotype groups also offers a simplified interpretation to be communicated to clinicians, who may find a system with several levels confusing and non-transparent.
The study included a large number of patients receiving vortioxetine treatment, which offers a unique insight into real-world clinical use of vortioxetine. To optimize personalized treatment with vortioxetine, other real-world PK data sets may be studied in the future. One of the limitations of the current study is that the data primarily originated from Caucasians. Evidence suggests that interactions between ethnicity and CYP2D6 genotype–phenotype relationships exist, and, furthermore, there are large interethnic differences in the frequency of CYP2D6 variant alleles. It would therefore be of great value in the future to study data sets originating from other ethnicities to better understand the impact of ethnicity.
Prediction of drug exposure from CYP2D6 genotypes and translation into dose recommendations is also challenged by the influence of other factors on drug metabolism. Although popPK modelling offers an opportunity to account for relevant patient factors when predicting exposure, it should be acknowledged that interpatient variability will exist in clinical practice regardless. Therefore, careful individual dose titration and routine TDM remain central to optimal personalized medicine, particularly within psychiatry.
5 Conclusion
The published popPK model of vortioxetine and its CYP2D6-mediated metabolite was externally validated using naturalistic patient data from TDM. The simulation- and prediction-based diagnostics showed that the model had a good ability to predict vortioxetine exposures observed in routine clinical practice, with a slight tendency to overpredict the vortioxetine concentrations. Different factors may have contributed to the prediction error, including poor treatment compliance among certain patients and, to a lesser extent, lack of information on patient characteristics and misspecified CYP2D6 alleles. The conventional CYP2D6 phenotype classification system (PM, IM, NM, UM) was shown to be adequate for predicting vortioxetine exposure.
References
Whiting B, Kelman AW, Grevel J. Population pharmacokinetics theory and clinical application. Clin Pharmacokinet. 1986;11:387–401.
Gaedigk A. Complexities of CYP2D6 gene analysis and interpretation. Int Rev Psychiatry. 2013;25:534–53.
Gaedigk A, Ingelman-Sundberg M, Miller NA, Leeder JS, Whirl-Carrillo M, Klein TE. The Pharmacogene Variation (PharmVar) Consortium: incorporation of the human cytochrome P450 (CYP) allele nomenclature database. Clin Pharmacol Ther. 2018;103:399–401.
Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS. The CYP2D6 Activity Score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther. 2008;83:234–42.
Gaedigk A, Dinh JC, Jeong H, Prasad B, Leeder JS. Ten Years’ experience with the CYP2D6 Activity Score: a perspective on future investigations to improve clinical predictions for precision therapeutics. J Pers Med. 2018;8:15.
Zhou SF. Polymorphism of human cytochrome P450 2D6 and its clinical significance: part II. Clin Pharmacokinet. 2009;48:761–804.
Thase ME, Mahableshwarkar AR, Dragheim M, Loft H, Vieta E. A meta-analysis of randomized, placebo-controlled trials of vortioxetine for the treatment of major depressive disorder in adults. Eur Neuropsychopharmacol. 2016;26:979–93.
Baldwin DS, Chrones L, Florea I, Nielsen R, Nomikos GG, Palo W, et al. The safety and tolerability of vortioxetine: analysis of data from randomized placebo-controlled trials and open-label extension studies. J Psychopharmacol. 2016;30:242–52.
Hiemke C, Bergemann N, Clement HW, Conca A, Deckert J, Domschke K, et al. Consensus guidelines for therapeutic drug monitoring in neuropsychopharmacology: update 2017. Pharmacopsychiatry. 2018;51:9–62.
Areberg J, Petersen KB, Chen G, Naik H. Population pharmacokinetic meta-analysis of vortioxetine in healthy individuals. Basic Clin Pharmacol Toxicol. 2014;115:552–9.
Hvenegaard MG, Bang-Andersen B, Pedersen H, Jorgensen M, Puschl A, Dalgaard L. Identification of the cytochrome P450 and other enzymes involved in the in vitro oxidative metabolism of a novel antidepressant, Lu AA21004. Drug Metab Dispos. 2012;40:1357–65.
Frederiksen T, Areberg J, Schmidt E, Bjerregaard Stage T, Brøsen K. Quantification of in vivo metabolic activity of CYP2D6 genotypes and alleles through population pharmacokinetic analysis of vortioxetine. Clin Pharmacol Ther. 2020;109:150–9.
Mannheimer B, Haslemo T, Lindh JD, Eliasson E, Molden E. Risperidone and venlafaxine metabolic ratios strongly predict a CYP2D6 poor metabolizing genotype. Ther Drug Monit. 2016;38:127–34.
Haslemo T, Eliasson E, Jukic MM, Ingelman-Sundberg M, Molden E. Significantly lower CYP2D6 metabolism measured as the O/N-desmethylvenlafaxine metabolic ratio in carriers of CYP2D6*41 versus CYP2D6*9 or CYP2D6*10: a study on therapeutic drug monitoring data from 1003 genotyped Scandinavian patients. Br J Clin Pharmacol. 2019;85:194–201.
Caudle KE, Sangkuhl K, Whirl-Carrillo M, Swen JJ, Haidar CE, Klein TE, et al. Standardizing CYP2D6 genotype to phenotype translation: consensus recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin Transl Sci. 2019;13:116–24.
Caudle KE, Dunnenberger HM, Freimuth RR, Peterson JF, Burlison JD, Whirl-Carrillo M, et al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet Med. 2017;19:215–23.
Glass PS, Shafer S, Reves JG. Intravenous drug delivery systems. Miller’s Anesth. Philadelphia: Elsevier; 2004. pp. 439–80.
Zhou Y, Ingelman-Sundberg M, Lauschke VM. Worldwide distribution of cytochrome P450 alleles: a meta-analysis of population-scale sequencing projects. Clin Pharmacol Ther. 2017;102:688–700.
US Drug label. TRINTELLIX (vortioxetine) tablets, for oral use. https://www.accessdata.fda.gov/drugsatfda_docs/label/2018/204447s017lbl.pdf. Accessed 30 Oct 2020.
DiMatteo MR, Lepper HS, Croghan TW. Depression Is a risk factor for noncompliance with medical treatment: meta-analysis of the effects of anxiety and depression on patient adherence. Arch Intern Med. 2000;160:2101–7.
Areberg J, Dragheim M. Assessment of patient compliance using pharmacokinetic data in clinical trials in depressed patients treated with vortioxetine [poster presentation]. British Association Psychopharmacology Summer Meeting 2015, University of Bristol.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
Trine Frederiksen received funding from Innovation Fund Denmark (grant number 8053-00083B) and H. Lundbeck A/S.
Conflict of interest
Trine Frederiksen and Johan Areberg are employed by H. Lundbeck A/S. Robert L. Smith, Birgit M. Wollmann, and Espen Molden have no conflicts of interest to declare.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
The datasets generated and/or analysed during the current study are not publicly available due to privacy issues but are available from the corresponding author upon reasonable request.
Code availability
Not applicable.
Author contributions
All authors contributed to the design of the study. Data collection and analysis was performed by RLS, BMW and TF. The first draft of the manuscript was written by TF and all authors had the possibility of commenting. The final version of the manuscript was read and approved by all authors.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Frederiksen, T., Smith, R.L., Wollmann, B.M. et al. Validation of a Population Pharmacokinetic Model of Vortioxetine Using Therapeutic Drug Monitoring Data. Clin Pharmacokinet 60, 1475–1486 (2021). https://doi.org/10.1007/s40262-021-01029-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40262-021-01029-7