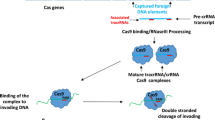
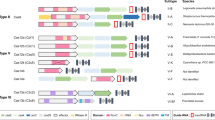
Abstract
The class 2 clustered regularly interspaced short palindromic repeats (CRISPR)-Cas system, one of the prokaryotic adaptive immune systems, has sparked a lot of attention for its use as a gene editing tool. Currently, type II, V, and VI effector modules of this class have been characterized and extensively tested for nucleic acid editing, imaging, and disease diagnostics. Due to the unique composition of their nuclease catalytic center, the effector modules substantially vary in their function and possible biotechnology applications. In this review, we discuss the structural and functional diversity in class 2 CRISPR effectors, and debate their suitability for nucleic acid targeting and their shortcomings as gene editing tools.






Similar content being viewed by others
References
Hille F, Richter H, Wong SP, Bratovič M, Ressel S, Charpentier E. The biology of CRISPR-Cas: backward and forward. Cell. 2018;172(6):1239–59.
Nuñez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna JA. Cas1–Cas2 complex formation mediates spacer acquisition during CRISPR–Cas adaptive immunity. Nat Struct Mol Biol. 2014;21(6):528.
Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78.
Makarova KS, Zhang F, Koonin EV. SnapShot: class 1 CRISPR-Cas systems. Cell. 2017;168(5):946.e1.
Makarova KS, Zhang F, Koonin EV. SnapShot: class 2 CRISPR-Cas systems. Cell. 2017;168(1):328.e1.
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23.
Knott GJ, Doudna JA. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361(6405):866–9.
Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, et al. Diversity and evolution of class 2 CRISPR–Cas systems. Nat Rev Microbiol. 2017;15(3):169.
Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, et al. An updated evolutionary classification of CRISPR–Cas systems. Nat Rev Microbiol. 2015;13(11):722.
Yan WX, Hunnewell P, Alfonse LE, Carte JM, Keston-Smith E, Sothiselvam S, et al. Functionally diverse type V CRISPR-Cas systems. Science. 2019;363(6422):88–91.
Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, Hsu PD. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173(3):665–676.e14.
Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60(3):385–97.
Smargon AA, Cox DB, Pyzocha NK, Zheng K, Slaymaker IM, Gootenberg JS, et al. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol Cell. 2017;65(4):618–630.e7.
Yan WX, Chong S, Zhang H, Makarova KS, Koonin EV, Cheng DR, et al. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol Cell. 2018;70(2):327–339.e5.
Zhang H, Dong C, Li L, Wasney GA, Min J. Structural insights into the modulatory role of the accessory protein WYL1 in the type VI-D CRISPR-Cas system. Nucleic Acids Res. 2019;47(10):5420–8.
Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156(5):935–49.
Nishimasu H, Cong L, Yan WX, Ran FA, Zetsche B, Li Y, et al. Crystal structure of Staphylococcus aureus Cas9. Cell. 2015;162(5):1113–26.
Fonfara I, Le Rhun A, Chylinski K, Makarova KS, Lecrivain A-L, Bzdrenga J, et al. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2013;42(4):2577–90.
Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343(6176):1247997.
Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–71.
Gao P, Yang H, Rajashankar KR, Huang Z, Patel DJ. Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition. Cell Res. 2016;26(8):901.
Dong D, Ren K, Qiu X, Zheng J, Guo M, Guan X, et al. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature. 2016;532(7600):522.
Yang H, Gao P, Rajashankar KR, Patel DJ. PAM-dependent target DNA recognition and cleavage by C2c1 CRISPR-Cas endonuclease. Cell. 2016;167(7):1814–1828.e12.
Liu J-J, Orlova N, Oakes BL, Ma E, Spinner HB, Baney KL, et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature. 2019;566(7743):218.
Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436–9.
Li S-Y, Cheng Q-X, Liu J-K, Nie X-Q, Zhao G-P, Wang J. CRISPR-Cas12a has both cis-and trans-cleavage activities on single-stranded DNA. Cell Res. 2018;28(4):491.
Swarts DC, Jinek M. Mechanistic Insights into the cis-and trans-acting DNase activities of Cas12a. Mol Cell. 2019;73(3):589–600.e4.
Harrington LB, Burstein D, Chen JS, Paez-Espino D, Ma E, Witte IP, et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362(6416):839–42.
Liu L, Li X, Wang J, Wang M, Chen P, Yin M, et al. Two distant catalytic sites are responsible for C2c2 RNase activities. Cell. 2017;168(1–2):121–134.e12.
Zhang B, Ye W, Ye Y, Zhou H, Saeed AF, Chen J, et al. Structural insights into Cas13b-guided CRISPR RNA maturation and recognition. Cell Res. 2018;28(12):1198.
Zhang C, Konermann S, Brideau NJ, Lotfy P, Wu X, Novick SJ, et al. Structural basis for the RNA-guided ribonuclease activity of CRISPR-Cas13d. Cell. 2018;175(1):212–223.e17.
Slaymaker IM, Mesa P, Kellner MJ, Kannan S, Brignole E, Koob J, et al. High-resolution structure of Cas13b and biochemical characterization of RNA targeting and cleavage. Cell Rep. 2019;26(13):3741–3751.e5.
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–21.
Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471(7340):602.
O’Connell M. Molecular mechanisms of RNA-Targeting by Cas13-containing type VI CRISPR-Cas systems. J Mol Biol. 2019;431(1):66–87.
Yin K, Gao C, Qiu J-L. Progress and prospects in plant genome editing. Nat Plants. 2017;3(8):17107.
Zhang Y, Long C, Li H, McAnally JR, Baskin KK, Shelton JM, et al. CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice. Sci Adv. 2017;3(4):e1602814.
Hu X, Wang C, Liu Q, Fu Y, Wang K. Targeted mutagenesis in rice using CRISPR-Cpf1 system. J Genet Genom. 2017;44(1):71–3.
O’connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516(7530):263.
Dugar G, Leenay RT, Eisenbart SK, Bischler T, Aul BU, Beisel CL, et al. CRISPR RNA-dependent binding and cleavage of endogenous RNAs by the Campylobacter jejuni Cas9. Mol Cell. 2018;69(5):893–905.e7.
Rousseau BA, Hou Z, Gramelspacher MJ, Zhang Y. Programmable RNA cleavage and recognition by a natural CRISPR-Cas9 system from Neisseria meningitidis. Mol Cell. 2018;69(5):906–914.e4.
Strutt SC, Torrez RM, Kaya E, Negrete OA, Doudna JA. RNA-dependent RNA targeting by CRISPR-Cas9. Elife. 2018;7:e32724.
Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, et al. RNA targeting with CRISPR–Cas13. Nature. 2017;550(7675):280.
Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353(6299):aaf5573.
Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–42.
Myhrvold C, Freije CA, Gootenberg JS, Abudayyeh OO, Metsky HC, Durbin AF, et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360(6387):444–8.
Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360(6387):439–44. https://doi.org/10.1126/science.aaq0179.
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–83.
Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–51.
Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, et al. Editing DNA methylation in the mammalian genome. Cell. 2016;167(1):233–247.e17.
Lei Y, Zhang X, Su J, Jeong M, Gundry MC, Huang Y-H, et al. Targeted DNA methylation in vivo using an engineered dCas9-MQ1 fusion protein. Nat Commun. 2017;8:16026.
Choudhury SR, Cui Y, Lubecka K, Stefanska B, Irudayaraj J. CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter. Oncotarget. 2016;7(29):46545.
Hilton IB, D’ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33(5):510.
Chen X, Wei M, Liu X, Song S, Wang L, Yang X, et al. Construction and validation of the CRISPR/dCas9-EZH2 system for targeted H3K27Me3 modification. Biochem Biophys Res Commun. 2019;511(2):246–52.
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420.
Cox DB, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, et al. RNA editing with CRISPR-Cas13. Science. 2017;358(6366):1019–27.
Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, et al. Programmable base editing of A• T to G• C in genomic DNA without DNA cleavage. Nature. 2017;551(7681):464.
Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li G-W, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155(7):1479–91.
Nelles DA, Fang MY, O’Connell MR, Xu JL, Markmiller SJ, Doudna JA, et al. Programmable RNA tracking in live cells with CRISPR/Cas9. Cell. 2016;165(2):488–96.
Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827.
Casini A, Olivieri M, Petris G, Montagna C, Reginato G, Maule G, et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat Biotechnol. 2018;36(3):265.
Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, et al. Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature. 2017;550(7676):407.
Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36(8):765.
Cullot G, Boutin J, Toutain J, Prat F, Pennamen P, Rooryck C, et al. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat Commun. 2019;10(1):1136.
Jin S, Zong Y, Gao Q, Zhu Z, Wang Y, Qin P et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019;364(6437):292–5.
Zuo E, Sun Y, Wei W, Yuan T, Ying W, Sun H et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019;364(6437):289–92.
Acknowledgements
We would like to acknowledge Lora Starrs and Alexander McKay for fruitful discussions and critical reading of the manuscript. Dr. Burgio’s research program is supported by the National Health and Medical Research Council of Australia (APP1143008), the Australian Research Council (DP180101494), and the National Collaborative Research Infrastructure Strategy (NCRIS) via the Australian Phenomics Network. Arash Hajizadeh Dastjerdi is supported by a JCSMR postgraduate scholarship, and Anthony Newman is supported by a Research Training Program domestic scholarship. We finally wish to acknowledge two anonymous reviewers and the editor for their helpful and excellent suggestions on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
No funding was received for the preparation of this review.
Conflict of interest
AHD, AN, and GB declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Hajizadeh Dastjerdi, A., Newman, A. & Burgio, G. The Expanding Class 2 CRISPR Toolbox: Diversity, Applicability, and Targeting Drawbacks. BioDrugs 33, 503–513 (2019). https://doi.org/10.1007/s40259-019-00369-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40259-019-00369-y