Abstract
Introduction
Aniridia is a rare congenital vision-loss disease caused by heterozygous variants in the PAX6 gene. There is no vision-saving therapy, but one exciting approach is to use CRISPR/Cas9 to permanently correct the causal genomic variants. Preclinical studies to develop such a therapy in animal models face the challenge of showing efficacy when binding human DNA. Thus, we hypothesized that a CRISPR gene therapy can be developed and optimized in humanized mouse embryonic stem cells (ESCs) that will be able to distinguish between an aniridia patient variant and nonvariant chromosome and lay the foundation for human therapy.
Methods
To answer the challenge of binding human DNA, we proposed the “CRISPR Humanized Minimally Mouse Models” (CHuMMMs) strategy. Thus, we minimally humanized Pax6 exon 9, the location of the most common aniridia variant c.718C > T. We generated and characterized a nonvariant CHuMMMs mouse, and a CHuMMMs cell-based disease model, in which we tested five CRISPR enzymes for therapeutic efficacy. We then delivered the therapy via lipid nanoparticles (LNPs) to alter a second variant in ex vivo cortical primary neurons.
Results
We successfully established a nonvariant CHuMMMs mouse and three novel CHuMMMs aniridia cell lines. We showed that humanization did not disrupt Pax6 function in vivo, as the mouse showed no ocular phenotype. We developed and optimized a CRISPR therapeutic strategy for aniridia in the in vitro system, and found that the base editor, ABE8e, had the highest correction of the patient variant at 76.8%. In the ex vivo system, the LNP-encapsulated ABE8e ribonucleoprotein (RNP) complex altered the second patient variant and rescued 24.8% Pax6 protein expression.
Conclusion
We demonstrated the usefulness of the CHuMMMs approach, and showed the first genomic editing by ABE8e encapsulated as an LNP-RNP. Furthermore, we laid the foundation for translation of the proposed CRISPR therapy to preclinical mouse studies and eventually patients with aniridia.
Similar content being viewed by others
Why carry out this study? |
PAX6 aniridia is a rare congenital disease, that typically leads to blindness, for which there is no vision-saving therapy. |
An exciting approach to therapy is to use CRISPR/Cas9 to permanently correct the causal genomic variants. |
What was the hypothesis of the study? |
The main hypothesis was that a CRISPR gene therapy can be developed and optimized in humanized mouse embryonic stem cells that will be able to distinguish between an aniridia patient variant and nonvariant chromosome and lay the foundation for further preclinical mouse studies. |
What were the study outcomes? |
The main outcome was an optimized CRISPR therapeutic strategy using the base editor ABE8e, which corrected the most common aniridia patient variant at 76.8% in vitro, and when delivered as an LNP-encapsulated RNP altered a second patient variant rescuing 24.8% of Pax6 protein expression. |
What has been learned from the study? |
We showed the first genomic editing by ABE8e encapsulated as an LNP-RNP. |
We provided support for the main hypothesis laying the foundation for the translation of our optimized CRISPR therapy for further preclinical mouse studies. |
Introduction
Congenital aniridia is a rare vision-loss disease characterized by the underdevelopment and malformation of the eye [1, 2]. Clinical features primarily include varying severities of hypoplasia of the iris, fovea, and optic nerve [1,2,3]. Patients experience low visual acuity and photophobia, which typically progressively worsens over time due to the occurrence of cornea keratopathy, glaucoma, and other disease manifestation, leading to blindness by young adulthood [3,4,5]. Aniridia is caused by more than 600 heterozygous pathogenic variants in the transcription factor paired box 6 (PAX6), a master regulator of ocular development, with dosage sensitivity in the eye [6,7,8,9]. The majority of these variants are dominant loss of function, and lead to phenotype due to PAX6 haploinsufficiency [10]. These include the most commonly reported aniridia patient variant, c.718C > T (p.R240X) located in exon 9 [11,12,13,14], which is one of four that together account for more than 20% of aniridia cases [7]. There are interventions that prolong vision [3] and ongoing work to develop drugs that may regulate PAX6 expression [15], but there are currently no vision-saving therapies for aniridia [16]. Thus, there is an unmet therapeutic need.
Fortunately, there has been considerable investigation and characterization of aniridia models to study molecular pathophysiology, disease progression, and therapeutic development [17]. Importantly, there is a therapeutic window, as demonstrated by the use of the nonsense suppressing drug, ataluren, in the small eye (Sey) aniridic mouse [18]. The Sey mouse presents with similar ocular phenotypes to those observed in patients with aniridia [19, 20] and is caused by a Pax6 nonsense variant, c.580G > T (p.G194X), which has also been reported in human [13]. Delivery of ataluren in juvenile aniridic mice positively improved phenotype, suggesting a therapeutic window in the early postnatal years in humans [15, 18], despite an unsuccessful clinical trial (NCT02647359). To further improve the Sey mouse model, we previously added a FLAG-tag to the Sey Pax6 allele (referred hereafter as Fey), which enables histological quantification of rescued Pax6 protein expression (MMRRC 066963-MU) [21].
One major challenge when undertaking therapy development with an animal model can be the requirement for preclinical studies to show efficacy on human DNA, RNA, or protein. This is particularly the case for the exciting CRISPR/Cas9-based approaches to therapy development, which require binding DNA [22]. To answer this challenge, we have developed the “CRISPR Humanized Minimally Mouse Models” (CHuMMMs) strategy. For the CHuMMMs strategy, we propose using CRISPR to engineer into the model a human-DNA “landing pad” to allow the CRISPR therapeutic reagents to bind human DNA at the site of the pathogenic variant. Whereas previous studies have shown that humanization of entire genes is technically demanding and costly and can have adverse consequences for gene function [23], the CHuMMMs strategy avoids these problems by humanizing only the minimal region needed for binding of the therapeutic CRISPR reagents. This approach will enable more rapid development of directly translatable CRISPR-based therapies.
CRISPR can establish targeted, permanent edits within the genome [24, 25]. Notably, the first in vivo CRISPR-based therapy for a congenital vision-loss disease, EDIT-101 (NCT03872479) [25, 26], demonstrated proof-of-concept and favorable safety profile across all dose cohorts in a phase I/II trial (https://ir.editasmedicine.com/press-releases, 25 January 2023). Since the discovery of this gene editing platform, there has been a great deal of development to improve upon the traditional CRISPR system [26]. Multiple orthologous wild-type (WT) Cas9 enzymes have been widely studied, while other researchers have engineered the WT Cas9 to ease protospacer adjacent motif (PAM) requirements, increase editing activity, and decrease off-target editing [27,28,29,30]. Beyond the traditional homology-directed repair (HDR) approach, which aims to exchange the pathogenic genomic sequence with a WT donor DNA template [31], new CRISPR systems have been engineered. These include base editors (BEs), which can confer selective single-base transition conversions [32]. In addition, BEs do not require donor DNA and do not produce double-stranded breaks (DSB) in genomic DNA, resulting in low rates of indels and less off-target editing [32, 33], which make them advantageous in developing clinical therapies. Overall, this provides many options to consider during therapy development. This rapid advancement of CRISPR technology suggests that personalized CRISPR therapies for low-frequency variants may be an effective approach to treat aniridia.
There are two primary delivery options for CRISPR-based therapies, recombinant adeno-associated viruses (rAAVs), and lipid nanoparticles (LNPs). rAAVs have been used to successfully deliver CRISPR components and produce genomic editing in multiple tissues, including the eye [34,35,36]. However, one major disadvantage of using rAAVs is their limited packaging capacity, ~ 4.9 kb [37]. With the most commonly used Cas9 enzyme, SpCas9, being ~ 4.1 kb, and a popular iteration of the adenine base editor, ABE8e [38], being ~ 4.8 kb in size, this leaves little space to encode additional components including: promoter, guide RNA (gRNA), and DNA template [34, 39, 40]. The second primary delivery method is to encapsulate CRISPR components in LNPs, which are less restricted in their packaging abilities and are scalable and non-immunogenic [41], therefore making the approach very attractive. Others have shown successful transfection and genomic editing using LNPs encapsulating SpCas9 and ABE8e mRNA [42,43,44,45]. However, less studied is the delivery with LNPs of the ribonucleoprotein (RNP) form of CRISPR, despite the fact that RNPs enable the use of the active form of the enzyme and chemically modified single-guide RNAs (sgRNAs), improving on-target editing and safety by minimizing off-target editing [46, 47].
Here we hypothesize that a CRISPR gene therapy can be developed and optimized in humanized mouse embryonic stem cells (ESCs) that will be able to distinguish between the patient variant and nonvariant chromosomes, thus laying the foundation for further preclinical studies correcting aniridic congenital blindness in mice. Having found support for this first hypothesis, a follow-up hypothesis was developed and tested, that our optimized CRISPR therapy could alter a second aniridia variant in a clinically relevant cell type via LNPs.
Methods
Isolation and Culture of Mouse ESCs
Male Pax6 WT C57BL/6NTac (Taconic, Hudson, NY) ESCs (mEMS6131 [48]) were derived as previously described [49], and cultured at 37 °C with 5% CO2 on either mouse embryonic fibroblasts (MEFs) or 0.1% gelatin. ESCs were maintained in ESC media and passaged as previously described [49].
RNP Design for Humanization of ESCs
Two gRNAs (cgEMS9, cgEMS18; Table S1, guide and template sequences) were designed to introduce two DSB in Pax6 (Fig. 1A). CRISPR RNAs (crRNA) and tracer RNAs (tracrRNA) were synthesized as single strands with chemical modifications (2′-O-methyl and phosphorothioate bonds at the first two 5′- and 3′-terminal RNA residues) (GenScript, Piscataway, NJ). A 512 bp single-stranded oligodeoxynucleotide (ssODN) template containing the patient variant (oEMS6346) was synthesized to confer the humanization of Pax6 exon 9, and an 84 bp ssODN (oEMS6451) was synthesized to confer correction of the variant in the resulting homozygous humanized variant cell line (Integrated DNA Technologies, Coralville, IA).
Derivation of homozygous humanized Pax6 exon-9 mouse ESCs, including an exon-9 pathogenic patient variant. A Schematic of humanization using a dual-gRNA strategy to exchange mouse genomic sequence with a 512 bp ssODN at Pax6 exon 9. ssODN consists of 100 bp mouse homology arms (blue) flanking 312 bp of human DNA sequence containing pathogenic patient variant, c.718C > T (red). cgEMS, CRISPR guide; oEMS, oligodeoxynucleotide; HA, homology arm. B Schematic of single-gRNA strategy to correct patient variant, c.718C > T, in humanized homozygous variant cell line, using an 84 bp ssODN to derive heterozygous and homozygous nonvariant cell lines. C Human-specific DNA amplified from single embryonic stem cell clones picked following CRISPR-based humanization of Pax6 exon 9. Lane 1, positive control DNA from a He9+/He9+ mouse ear notch. Lane 2, DNA from a clone positive for humanization event (mEMS6634). Lane 3, negative control DNA from a B6 WT mouse. Lane 4, no template negative control. Lanes 5 and 6, DNA from clones positive for the humanization event (mEMS6670 and mEMS6674, respectively). Band at 265 bp indicates humanization. D Restriction fragment length polymorphism screen of the PCR products shown in B using AvaI restriction enzyme identifies He9−/He9− clones that have undergone either heterozygous (lane 5) or homozygous (lane 6) CRISPR correction of the patient variant. Fragment lengths of 145 bp and 117 bp indicate DNA cut by AvaI due to the presence of the nonvariant base. Fragment length of 265 bp indicate uncut DNA due to presence of variant. E Sanger sequencing showing location of patient variant (red) from humanized cell lines demonstrating homozygosity (mEMS6634) or heterozygosity (mEMS6670) for patient variant, or homozygosity (mEMS6674) for nonvariant sequence. He9, humanized exon 9
Cell Transfection and Picking Single Clones
ESCs were passaged in a 1:2 split ratio 24 h prior to transfection and fed with fresh media 2 h prior to transfection. Cells were dissociated using Trypsin–EDTA (catalog 25,200–072, Invitrogen, Thermo Fisher) and counted using a hemocytometer. gRNAs were prepared by annealing crRNA and tracrRNA at 95 °C for 5 min. RNP was prepared by complexing SpCas9 protein with each gRNA for 15 min at room temperature (RT), prior to the addition of the ssODN and WT mouse ESCs for transfection. Each reaction consisted of 9 µL ESCs (2 × 106 per reaction) mixed with 0.3 µL of RNP and 0.2 µL of 5 µM ssODN and was electroporated using Neon Transfection System (catalog MPK5000, Invitrogen) on setting 14 (1200 mV, width 20, Pulse #2). Electroporated ESCs were plated onto a fresh 24-well plate on either MEFs or gelatin and incubated for 48 h at 37 °C with 5% CO2. Cells were either harvested for molecular characterization or cryopreserved, as previously described [49]. For ESC clones, electroporated cells were thawed and plated in serial dilutions on 0.1% gelatin in 6 cm dishes and incubated for 48 h at 37 °C with 5% CO2. Individual clones were isolated and plated on 96-well gelatinized plates and incubated at 37 °C with 5% CO2 until > 80% confluent.
DNA Isolation, PCR Genotyping, and RFLP Screens
Once ESCs became confluent, cells were digested in tissue homogenization buffer with Proteinase K according to a previously described protocol [49]. DNA from lysed cell samples was amplified using Taq DNA Polymerase (catalog 18,038,042, Invitrogen) and PCR primer pairs (Table S2, primer sequences) specifically targeting: human sequence inside the humanized region, 5′ mouse–human junction, 3′ human–mouse junction, 5′ mouse–3′ mouse flanking the humanized region, and mouse–mouse inside of the region that was humanized, were used to confirm successful humanization of Pax6 exon 9 (Fig. 1A). Candidates for successful humanization were confirmed by Sanger sequencing.
To screen for heterozygous correction of patient variant c.718C > T, a restriction fragment length polymorphism (RFLP) assay using AvaI (catalog R0152S, NEB) and DdeI (catalog R0175S, NEB) restriction enzymes was used. DNA was first amplified with oEMS6223 and oEMS6224 before incubation with CutSmart Buffer (catalog B6004S, NEB) and either restriction enzyme at 37 °C for 1 h, then inactivation at either 80 °C or 65 °C for 10 min. AvaI cuts this DNA in presence of the cytosine base and DdeI cuts in presence of the thymine base, giving rise to fragments 117 bp and 145 bp. Candidates for heterozygous and homozygous correction of patient variant were confirmed by sequencing.
Sanger Sequencing and Peak Quantification
The region around the humanized exon (813 bp) was PCR amplified with appropriate primer pairs (Table S2). PCR products were run on a 2% agarose gel for 40 min at 130 V. Bands were excised and DNA was purified using QIAquick Gel Extraction Kit (catalog 28,706, QIAGEN, Germantown, MD). Bidirectional sequencing was carried out by the CMMT DNA Sequencing Core Facility and the UBC Sequencing and Bioinformatics Consortium. Chromatograms were viewed using Benchling (www.benchling.com).
For CRISPR therapy optimization experiments, we sequenced unidirectionally in the reverse direction. In this direction, the target bases were sequenced prior to the cut site of Cas9. Peak height data were extracted from CRISPR-treated and mock (untreated) samples using Analysis Module Variant Analysis (VA) software (catalog A28220, Thermo Fisher) and EditR software [50]. The treated peak height data were normalized by subtracting the average of mock untreated He9−/He9− replicas to remove background and conservatively calculate CRISPR editing. No samples were excluded from analysis.
Generation of Humanized Exon 9 Mice
Mouse strains were derived by means of cytoplasmic microinjection into C57BL/6J mice (JAX 000,664) using a dual RNA guide strategy (cgEMS9, cgEMS18; Table S1) and ssODN (oEMS6347) according to a previously described protocol [21].
Initially, two mouse strains were derived from two independent founders, C57BL/6J-Pax6em6(PAX6)Ems (MGI: 7,330,073) and C57BL/6J-Pax6em7(PAX6)Ems (MGI: 7,330,075). They were indistinguishable by casual observation, so the studies conducted here used C57BL/6J-Pax6em7(PAX6)Ems, which for clarity and brevity will be called humanized Pax6 exon 9 nonvariant or B6-He9+. A third strain was then derived, 129S1.B6- Pax6em7(PAX6)Ems (MGI: 7,330,077) by backcrossing B6-He9+ onto the 129S1/SvImJ (JAX 002,448) genetic background, which for clarity and brevity will be called 129-He9+.
Phenotyping by Visual Inspection and Slit Lamp Imaging
B6-He9+/+ heterozygous N6 and N7 (backcrossed six and seven times) mice were mated in trios to produce He9+/He9+, He9+/+ , and +/+ offspring. This breeding scheme was also repeated mating 129-He9+/+ N6 trios to produce He9+/He9+, He9+/+, and +/+ offspring. External ocular morphology of adolescent (3–4 weeks) He9+/He9+ and +/+ mice was assessed by visual inspection and scored as either normal or abnormal. Adult (~ 2 months old) He9+/He9+ and +/+ mice were anesthetized using isoflurane at a flow rate of 1.5–1.8% in an induction chamber using a SomnoSuite (Kent Scientific, Torrington, CA). Once a surgical plane of anesthesia was induced, mice were transferred to the nose cone and eyes were covered with 1% Isopto Tears ophthalmic solution (ALCON, Geneva, Switzerland). Micron IV Retinal Imaging Microscope (Phoenix Research Labs, Pleasanton, CA) with an anterior segment slit lamp attachment was used to image the left eyes.
Purification of ABE8e Protein
CRISPR ABE8e protein was isolated by plasmid overexpression and purification [51]. pABE8e-protein plasmid (a gift from David Liu, #161,788, Addgene, Watertown, MA).
RNP Design and Complexation for CRISPR Therapy Development
sgRNAs (20 bp) and ssODNs (80 bp) were designed for CRISPR HDR and ABE8e. Chemical modifications of reagents were the same as above. sgRNAs and CRISPR enzymes were complexed for 15 min at RT prior to additional of ssODN (in the case of CRISPR HDR strategies) and ESCs for electroporation of two biological replicates per treatment group. ESCs were harvested for characterization by sequencing, as described above.
Preparation of Incisive Delivery System LNPs and Encapsulation of RNPs and DNA Template
Preparation of Incisive Delivery System LNPs was performed according to a previously described protocol [52]. The size distribution and the polydispersity index (PDI) of LNPs encapsulating ABE8e were measured using a Malvern Zetasizer Nano S instrument (Worcestershire, UK) (He–Ne laser, λ = 632 nm, detection angle 173°).
Ex Vivo Genome Editing
Treatment of Fey primary embryonic cortical neurons with LNP-encapsulated CRISPR-RNPs and subsequent analyses (immunocytochemistry, stereology, microscopy, and image processing) were performed according to a previously described protocol [52]. Minor changes were that treatments took place on day ex vivo (DEV) 6, and cells were collected on DEV 9.
Statistical Analysis
All statistics and plotting of graphs were conducted using GraphPad Prism version 9.4.0 for Windows (GraphPad Software, San Diego, CA). Statistical significance was determined using two-tailed unpaired t-test or Fisher’s exact test for comparisons of two groups, and one-way or two-way ANOVA analysis for three or more groups, where appropriate. One-way ANOVA tests were corrected for using Tukey’s multiple comparisons tests, while two-way ANOVA tests were corrected for using Sidak’s multiple comparisons tests. p value ≤ 0.05 was considered to be statistically significant.
Compliance with Ethics Guidelines
All animals were housed and bred in the pathogen-free Transgenic Animal facility at the Centre for Molecular Medicine and Therapeutics (CMMT) of the University of British Columbia (UBC). All mouse work was performed following protocols approved by the UBC Animal Care Committee (protocol numbers A21-0410, A21-0184), in accordance with guidelines determined by the Canadian Council on Animal Care.
Results
CHuMMMs for Pax6-Aniridia Therapy Development In Vitro
To implement the CHuMMMs strategy for aniridia in vitro we developed minimally humanized Pax6 ESC lines. Mouse ESCs were chosen for their ease of genetic manipulation, their differentiation potential, and ability to derive new mouse strains [21, 53,54,55]. Initially, we generated two novel homozygous minimally humanized exon-9 PAX6 variant c.718C > T (He9−/He9−) ESC lines (mEMS6634 and mEMS6658). A dual-gRNA strategy enabled the HDR-mediated humanization in WT C57BL/6N mouse ESCs by electroporation of CRISPR RNPs and an ssODN template (Fig. 1A and B) (Table S1, guide and template sequences; see electronic supplementary material). The sgRNAs introduced DSBs in Pax6 introns 8 and 9, respectively. A 512 bp ssODN containing the patient variant, c.718C > T, conferred the exchange of a 312 bp region of mouse DNA sequence with human sequence, via flanking 100 bp mouse-specific homology arms (HAs). Single clones were picked from electroporated cells. PCR assays confirmed successful insertion of the ssODN (Fig. 1C), and a RFLP screen determined that clones were either heterozygous or homozygous for the HDR event (Fig. 1D). Final confirmation of the 1.2 kb region (including 250 bp regions directly 5′ and 3′ of the ssODN) was completed by Sanger sequencing (Fig. 1E).
Subsequently, we generated one novel heterozygous minimally humanized exon-9 cell line (He9−/He9+; mEMS6670) and two humanized homozygous nonvariant cell lines (He9+/He9+; mEMS6674 and mEMS6676) to complete this cell-based disease model (Fig. 1B). A sgRNA (Table S1) and an 84 bp ssODN were employed to correct the patient variant in the He9−/He9− cell line mEMS6634 derived above. Single clones were again picked, and successful correction of the patient variant was characterized by PCR assays (Fig. 1C), the RFLP screen (Fig. 1D), and sequencing of the 1.2 kb region (Fig. 1E).
Minimal Humanization with Nonvariant PAX6 Results in No Phenotype In Vivo
Prior to therapy development using the CHuMMMs cell lines, we wanted to ensure that the minimal humanization alone did not cause adverse consequences for Pax6 gene function. Since there are no amino acid differences between mouse and human this seemed likely; however, changes in codon usage, splicing sites, and potential transcription binding sites were concerns. We reasoned that the ultimate determinant of Pax6 gene function was eye development in vivo.
Thus, we established CHuMMMs He9+/ + mice through direct injection of CRISPR reagents, using the same dual-gRNA strategy described above, and the nonvariant ssODN (Table S1). The resulting founder mice were made and bred on the C57BL/6J (B6) genetic background. The fully characterized strain was C57BL/6J-Pax6em7(PAX6)Ems (called B6-He9+ hereafter). Sequencing of the 1.2 kb exon-9 region described above confirmed the successful molecular event. The B6-He9+ strain was then bred onto a 129S1/SvImJ (129) genetic background to derive 129S1/SvImJ-Pax6em7(PAX6)Ems (called 129-He9+ hereafter).
The observed normal ocular morphology of He9+/He9+ mice gave evidence that the minimal humanization event at exon 9 alone did not disrupt Pax6 gene function in vivo. We characterized mice by visual inspection and found that the external ocular morphology of B6-He9+/He9+ mice did not differ significantly from B6 WT mice. External ocular morphology of B6-He9+/He9+ mice did, however, differ from the well-characterized B6-Sey/+ mouse, as shown by slit lamp images (Fig. 2A). This work was repeated with 129-He9+/He9+ and 129 WT mice, and again we observed no significant difference between the two groups (Fig. 2B).
CRISPR-based minimal humanization of Pax6 does not result in an ocular phenotype in mouse. A and B Slit lamp images of WT He9+/He9+ and Sey/+ mice demonstrated that He9+/He9+ ocular phenotype does not differ from WT and does differ from the Sey/+ mice aniridia phenotype. Mice were phenotyped by visual inspection and eyes scored as either “normal” or “abnormal.” A The He9+ allele was bred onto a C57BL/6J background and compared to C57BL/6J WT controls (He9+/He9+, n = 84; WT, n = 90). B The He9+ allele was bred onto a 129S1/SvImJ background and was compared with 129S1/SvImJ WT controls (He9+/He9+, n = 87; WT, n = 94). ns, no significant difference
SpCas9 Gave Higher Editing of Patient Variant than SaCas9
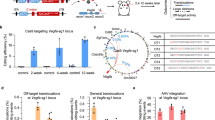
To compare SpCas9 versus SaCas9 we used He9−/He9− and He9+/He9+ ESCs. The He9−/He9− cells allowed for the observation and quantification of correction of the patient variant, and the He9+/He9+ cells allowed for the observation and quantification of unwanted alteration on the nonvariant chromosome (Fig. 3). For SpCas9, a single optimal sgRNA and ssODN were chosen. For SaCas9 a single optimal sgRNA and two ssODNs were chosen (Table S1). In addition to the correction of the c.718C > T, the ssODNs included synonymous single base mismatches, referred to here as “blocking mutations,” to prevent repeated targeting by Cas9 of edited alleles. These blocking mutations also enabled the quantification of on-target alteration on the nonvariant chromosome. RNPs were delivered by electroporation, including replicas, and total cell lysates were harvested for characterization. To quantify CRISPR editing by Sanger sequencing, we focused our analyses on unidirectional sequencing in which the target base was prior to, and unaffected by, indels at the Cas9 cut site. Peak height was normalized by subtracting the average of mock untreated He9−/He9− replicas to remove background and conservatively calculate the CRISPR editing.
CRISPR genome editing of the patient variant is higher using SpCas9 than SaCas9. A Top panel: Sanger sequencing of untreated He9−/He9− ESCs where patient variant c.718C > T (overlined) was homozygous for thymine as indicated by a singular red peak. Bottom panel: Sanger sequencing of SpCas9 treated He9−/He9− ESCs demonstrated correction of the patient variant from T to C at 34.3%, as shown by dominant red peak and secondary blue peak. The ssODN also included a synonymous “blocking mutation” (overlined) to prevent additional targeting of previously edited alleles. The blocking mutation was edited from T to C at 25.6%. PAM highlighted in red text. gRNA location shown by underlined text. B Left panel: Quantification of editing at the patient variant in He9−/He9− by SpCas9 was the highest average correction at 31.7 ± 3.8%. This was significantly different from the average editing by SaCas9 with oEMS6453 or oEMS6454, at 11.5 ± 6.3% and 9.37 ± 2.3%, respectively. Right panel: Quantification of editing at the site of the blocking mutation on the nonvariant chromosome in He9+/He9+ cells was not significantly different among the three strategies. This average editing by SpCas9 was at 4.02 ± 3.6%, by SaCas9 with oEMS6453 was at 2.14 ± 3.5%, and SaCas9 with oEMS6454 was at 0.460 ± 2.7%. ns, p > 0.05; *, p ≤ 0.05
We found that SpCas9 delivered to He9−/He9− ESCs showed significantly superior average editing of the patient variant from T to C at 31.7 ± 3.8% (Fig. 3A), in comparison to the average editing observed by SaCas9 with either ssODN at 11.5 ± 6.3% or 9.37 ± 2.3% (Fig. 3B). The results also showed that the average editing of the blocking mutation on the nonvariant chromosome in the He9+/He9+ cell line by SpCas9 at 4.02 ± 3.6%, and SaCas9 with either ssODN at 2.14 ± 3.5% or 0.460 ± 2.7%, was not significantly different among the three groups. Thus, SpCas9 was selected for further optimization in the subsequent experiments based on high editing of the patient variant and minimal editing on the nonvariant chromosome.
High-Fidelity Cas9 Gave Lower Alteration of Nonvariant Chromosome than WT SpCas9
To further optimize our CRISPR strategy, we compared average editing of three Cas9 nucleases from different commercial retailers: WT SpCas9, PNABio, Thousand Oaks, CA; “TrueCut” WT SpCas9, Invitrogen, Waltham, MA; and “high-fidelity” (HiFi) mutated SpCas9, Integrated DNA Technologies, Coralville, IA (Fig. 4). Experimental design was the same as described above for SpCas9 versus SaCas9.
A high-fidelity SpCas9 resulted in lower CRISPR editing on the nonvariant chromosome. A Left panel: Quantification of editing of the patient variant c.718C > T (overlined) in He9−/He9− cells found no significant difference between PNA Bio Cas9 and TrueCut Cas9 nucleases. Average editing observed by PNA Bio Cas9 was at 26.8 ± 6.0% and TrueCut Cas9 was at 34.9 ± 0.62%. Right panel: Quantification of editing of the synonymous blocking mutation (overlined) on the nonvariant chromosome in He9+/He9+ cells did not show a significant difference between the two nucleases. The average editing by PNA Bio Cas9 was at 13.9 ± 4.9% and TrueCut Cas9 was at 13.3 ± 3.2%. B Left panel: Quantification of editing of the patient variant in He9−/He9− cells found no significant difference between PNA Bio Cas9 and HiFi Cas9. Average editing observed by PNA Bio Cas9 was at 37.1 ± 2.7% and HiFi Cas9 was at 26.7 ± 4.7%. Right panel: Quantification of editing of the blocking mutation on the nonvariant chromosome in He9+/He9+ cells found HiFi Cas9 showed significantly lower editing than PNA Bio Cas9. The average editing by PNA Bio Cas9 was at 23.6 ± 1.9%, and HiFi Cas9 was at 0.350 ± 0.028%. C Left panel: Sanger sequencing of PNA Bio Cas9 treated He9+/He9+ cells showed a secondary blue peak at the blocking mutation demonstrating impact on the nonvariant chromosome. Also, secondary sequence is evident throughout the chromatogram 3′ of Cas9 cut site due to CRISPR-based insertions and deletions (indels). Right panel: Sanger sequencing of HiFi Cas9-treated He9+/He9+ cells demonstrated minimal CRISPR-based editing at the blocking mutation on the nonvariant chromosome and minimal secondary sequence throughout chromatogram due to indels. PAM highlighted in red text. gRNA location shown by underlined text. ns, p > 0.05; **, p ≤ 0.01
First, we studied the two WT SpCas9 enzymes (Fig. 4A). Average editing of the patient variant by PNABio SpCas9 at 26.8 ± 6.0% and TrueCut SpCas9 at 34.9 ± 0.62% was not significantly different. Average editing of the blocking mutation on the nonvariant chromosome by PNABio Cas9 at 13.9 ± 4.9% and TrueCut Cas9 at 13.3 ± 3.2% was also not significantly different. These results demonstrated that possible variabilities in manufacturing of nucleases by these commercial retailers did not affect average editing activity of the WT SpCas9.
We then tested the WT SpCas9 from PNA Bio, versus HiFi Cas9 (Fig. 4B and C). HiFi Cas9 has been engineered to reduce off-target editing while maintaining on-target editing through the introduction of a single point mutation in the REC1 sgRNA recognition domain to enhance specificity of sgRNA binding [56]. Average editing of the patient variant by PNABio Cas9 at 37.1 ± 2.7% and HiFi Cas9 at 26.7 ± 4.7% were not significantly different. Conversely, average editing of the blocking mutation on the nonvariant chromosome by PNABio Cas9 at 23.6 ± 1.9% and HiFi Cas9 at 0.350 ± 0.028% were significantly different. On the basis of these results, we found that the engineered HiFi Cas9 functioned in our assay as expected, and was the best SpCas9 choice for further HDR-mediated CRISPR therapy development.
ABE8e Gave Superior Editing of Patient Variant and Reduced Alteration of Nonvariant Chromosome than SpCas9
Next, we compared average editing of the patient variant by base editing using ABE8e versus HDR-mediated editing using WT SpCas9 (Fig. 5), following the same experimental design described above for SpCas9 versus SaCas9. As an ssODN is not required for ABE8e editing, we quantify on-target alteration on the nonvariant chromosome by a bystander edit observed 4 bp from the target base location. We tested two sgRNAs with ABE8e: cgEMS25 (Table S1) targeted an optimal PAM (NGG), and placed the target base at position 8 of the reported optimal editing window of positions 4–8 for ABE8e [38]; and cgEMS46 targeted a suboptimal PAM (NGA), but placed the target base at position 7 in the editing window, slightly more central to the optimal editing window, which is reported to improve on-target editing by ABE8e [38]. Throughout this study, thus far, standard chemical modifications of sgRNAs have been used [47]. Here, we also test unmodified sgRNAs, given that the success of using modified or unmodified sgRNAs will affect the choice of delivery methods.
ABE8e with a modified guide showed superior editing of the patient variant, with minimal impact on the nonvariant chromosome. A Left panel: Sanger sequencing of SpCas9-treated He9−/He9− cells demonstrated CRISPR-based editing of the patient variant c.718C > T (overlined) from T to C at 57.7%, as shown by dominant blue peak and secondary red peak. Right panel: Sanger sequencing of treated He9−/He9− cells with ABE8e complexed with cgEMS25 demonstrated highest CRISPR-based editing of the patient variant at 74.5%. Secondary peak observed 4 bp 5′ of the intended edit is due to synonymous bystander editing (overlined) by ABE8e. PAM highlighted in red text. gRNA location shown by underlined text. B Left panel: Quantification of average editing of patient variant in He9−/He9− cells showed treatment with SpCas9 resulted in editing at 41.8 ± 5.2%, ABE8e complexed with cgEMS25 showed significantly higher average editing at 73.0 ± 0.64%, and ABE8e with cgEMS46 showed significantly lower average editing at 6.37 ± 0.79%. Right panel: Treatment in He9+/He9+ cells resulted in average editing of the synonymous blocking mutation (overlined) on the nonvariant chromosome by SpCas9 at 15.4 ± 2.0%. Impact on the nonvariant chromosome by ABE8e was assayed by the alteration of the bystander edit. ABE8e with cgEMS25 resulted in a bystander edit at 0.545 ± 0.73%, and ABE8e with cgEMS46 resulted in a bystander edit at 0.390 ± 0.52% on the nonvariant chromosome, both of which show significantly lower average editing than SpCas9. C Left panel: Quantification of ABE8e complexed with chemically modified cgEMS25 in He9−/He9− cells showed significantly higher average editing than unmodified at 76.8 ± 0.48% and 2.87 ± 0.042%, respectively. ABE8e complexed with chemically modified cgEMS46 demonstrated significantly higher average editing than unmodified cgEMS46 at 3.82 ± 1.2% and 1.37 ± 0.14%, respectively. ns, p > 0.05; *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001
We found that ABE8e complexed with cgEMS25 showed significantly higher average editing of the patient variant at 73.0 ± 0.644% than ABE8e complexed with cgEMS46 at 6.37 ± 0.79%, and SpCas9 complexed with cgEMS25 at 41.8 ± 5.2% (Fig. 5A–B). These results demonstrate the superior editing activity of the patient variant by ABE8e versus WT SpCas9, as well as the importance of targeting the sgRNA to an optimal PAM. Additionally, positioning the target base more central to the editing window of ABE8e by 1 bp did not sufficiently improve editing activity, when paired with suboptimal PAM recognition by the sgRNA. Excitingly, ABE8e also showed significantly lower editing of the nonvariant chromosome than SpCas9 (Fig. 5B). ABE8e complexed with cgEMS25 resulted in an average bystander edit on the nonvariant chromosome at 0.545 ± 0.73% and ABE8e complexed with cgEMS46 resulted in an average bystander edit at 0.390 ± 0.52%, both of which showed significantly lower average editing of the nonvariant chromosome than SpCas9 at 15.4 ± 2.0%. These results demonstrate the superior ability of ABE8e to differentiate the single base pair difference between the patient variant and nonvariant chromosome at this locus, in comparison with the HDR-mediated approach.
To further optimize our ABE8e therapeutic strategy, we compared the use of chemically modified versus unmodified sgRNAs (Fig. 5C). We found that the average editing efficiency of the patient variant by ABE8e complexed with chemically modified cgEMS25 at 76.8 ± 0.48% was significantly higher than ABE8e complexed with unmodified cgEMS25 at 2.87 ± 0.042%. We also found that the average editing efficiency by ABE8e complexed with modified cgEMS46 at 3.82 ± 1.2% was significantly higher than ABE8e complexed with unmodified cgEMS46 at 1.37 ± 0.14%. Overall, these results confirm the importance of chemically modified sgRNAs to editing activity by ABE8e-RNPs.
ABE8e Edits Patient Variant “Additively” in Humanized Heterozygous Variant Cell Line
To further investigate how our ABE8e-mediated therapy would behave in patient cells, we compared the editing activity of the optimized ABE8e-mediated therapy in all three of the humanized cell lines (He9−/He9−, He9−/He9+, and He9+/He9+) to determine whether the patient variant would be edited additively or synergistically in the heterozygous cell line (Fig. 6). These results may give insight into how ABE8e would behave in aniridia patient cells, which are heterozygous for the pathogenic variant. The experimental design is the same as described above for SpCas9 versus SaCas9.
ABE8e edits patient variant “additively” in heterozygous variant cells. A Quantification of total % cytosine at site of patient variant c.718C > T following ABE8e treatment of He9−/He9+ cells was intermediate to, but not significantly different from, homozygous He9−/He9− and He9+/He9+ cells at 74.8 ± 5.0%, 65.3 ± 5.7%, and 89.8 ± 0.60%, respectively. B Quantification of the change in % cytosine at site of patient variant following ABE8e treatment of He9−/He9+ cells was intermediate to, and significantly different from, homozygous He9−/He9− and He9+/He9+ cells at 30.8 ± 5.0%, 65.3 ± 5.7%, and −1.98 ± 0.60%, respectively. ns, p > 0.05; *, p ≤ 0.05; **, p ≤ 0.01
We measured the total % cytosine (Total %C) at the site of the patient variant in the ABE8e-treated He9−/He9−, He9−/He9+, and He9+/He9+ cell lines and found that average editing of the variant in the He9−/He9+ cell line at 74.8 ± 5.0% was intermediate to, but not significantly different from, treatment in the He9−/He9− cells at 65.3 ± 5.7% and He9+/He9+ cells at 89.8 ± 0.60% (Fig. 6A). Here, typical variation and normalization by subtracting the average of mock untreated replicas reduced the He9+/He9+ peak value from the expected 100%. To further demonstrate the effect of the ABE8e treatment in each of these cell lines, we present the data again but now as a change in % cytosine (Delta %C) (Fig. 6B). We normalized ABE8e treated samples to %C at the site of the patient variant in the mock untreated samples, this time separately for each cell line. We found that the Delta %C in ABE8e treated He9−/He9+ cells at 30.8 ± 5.0% was intermediate to, and significantly different from, the Delta %C in ABE8e treated He9−/He9− cells at 65.3 ± 5.7% and He9+/He9+ cells at −1.98 ± 0.60%. These results demonstrate that ABE8e edited the patient variant additively in the heterozygous cell line, with no evidence of effect of the nonvariant chromosome.
LNP-Encapsulated ABE8e-RNPs Edited a Pax6 Patient Variant in Mouse Ex Vivo Cortical Neurons
As electroporation is not a translatable delivery method, we consider progressing development of our CRISPR therapy for aniridia to either rAAV or LNPs. Having demonstrated the importance of chemically modified sgRNAs to editing activity, which cannot be maintained when the sgRNA is encoded by rAAV, we decided to encapsulate ABE8e-RNPs in LNPs. For this we chose the Incisive Delivery System (Incisive Genetics Inc., Vancouver, Canada) for CRISPR-based gene therapies, which we have used successfully before to efficiently deliver SpCas9 RNP in vivo [52]. In addition, we moved the study from ESCs to the more clinically relevant mouse primary cortical neurons. Since the patient variant c.718C > T present in the He9− cells was not available in vivo, we shifted to another patient variant c.580G > T present in the Sey mouse. Thus, embryonic primary cortical neurons were derived from Fey mice. The 3xFLAG tag enabled histological quantification of correction of the Sey variant, by rescued Pax6 protein expression [21, 52]. This previously reported method found success using CRISPR SpCas9 HDR to correct the Sey variant to the WT guanine base. Here, we test ABE8e, which altered the variant thymine to a cytosine base, converting a stop codon to an arginine missense mutation. As such, we were initially uncertain whether this missense alteration would result in a stable Pax6 protein and, thus, detectable FLAG expression. We assayed successful transfection and genomic editing by Sanger sequencing and stereological analysis.
Excitingly, we found that ABE8e was successfully encapsulated by LNPs, transfected primary cortical neurons, altered the Sey variant at the genomic level, and rescued Pax6 protein expression. Transfection was first demonstrated by positive control SpCas9 HDR-treated cells showing FLAG expression, which colabeled with PAX6, indicative of correction of the patient variant to WT (Fig. 7A). FLAG expression and colabeling was also seen by the ABE8e (100 nM)-treated cells, indicative of alteration of the patient variant despite a perhaps less stable, but certainly detectable, missense-carrying Pax6 protein.
LNP delivered ABE8e RNP edited Pax6 pathogenic patient variant in mouse cortical neurons ex vivo. A Immunocytochemical images of 3xFLAG-tagged Pax6 Sey (Fey) embryonic mouse cortical neurons. Hoechst (blue) was used to reveal cell nuclei. As expected for embryonic cortical neurons, anti-PAX6 (red) showed positive cells in all samples, including the heterozygous Fey cells. Anti-FLAG (green) showed successful editing of Sey variant c.580G > T leading to expression of the corrected PAX6 protein in SpCas9 positive control and ABE8e (100 nM) RNP treated cells. Thus, as expected, anti-PAX6 and anti-FLAG showed colabeling in merge images (yellow). FLAG expression was very low in the ABE8e (50 nM)-treated cells and not observed in the untreated or negative control; the latter was SpCas9-RNP complexed with a luciferase-targeting guide. A further negative control was samples processed with no primary antibody. Images were taken at 20× magnification. Scale bar, 50 µm. B Quantification of editing of Sey variant in Fey ex vivo cortical neurons. ABE8e (100 nM)-treated cells showed significantly higher alteration of target variant compared with ABE8e (50 nM) and the luciferase negative control at 2.33 ± 1.0%, 0.340 ± 0.017%, and 0.00 ± 0.58%, respectively. C Quantification of FLAG-tagged Pax6 protein expression in Fey ex vivo cortical neurons. FLAG expression was significantly different between ABE8e (50 nM), ABE8e (100 nM), and the luciferase negative control treated groups at 6.83 ± 1.6%, 24.8 ± 1.3%, and 0.00 ± 0.0%, respectively. ns, p > 0.05; *, p ≤ 0.05; ***, p ≤ 0.001; ****, p ≤ 0.0001
Quantification by Sanger sequencing of whole cell lysates found that the ABE8e (100 nM)-treated cells showed a genomic alteration of the Sey variant at an average of 2.33 ± 1.0%, which was significantly greater than the alteration observed in the ABE8e (50 nM) at 0.340 ± 0.017% and luciferase control group at 0.00 ± 0.58% (Fig. 7B). The ABE8e (50 nM)-treated group was not significantly different from the negative control. Stereological quantification of protein expression showed that for the ABE8e (100 nM)-treated cells, FLAG expression, as a percentage of total Pax6 protein-expressing cells, at 24.8 ± 1.3% was significantly greater than that in both the ABE8e (50 nM)-treated cells at 6.83 ± 1.6% and the luciferase-targeting negative control group at 0.00 ± 0.0% (Fig. 7C). Thus, we can conclude that the higher molar concentration of ABE8e encapsulated in LNPs was more successful in editing the Sey patient variant to rescue Pax6 expression.
We suggest that the observed lower editing assayed by sequencing, compared with stereology, is a consequence of the large number of condensed nuclei (non-Pax6-expressing cells) present across all treatment groups (Fig. S1) [52]. Samples sent for sequencing were total cell lysates, and thus the non-Pax6-expressing neurons, would have diluted the result of ABE8e-mediated editing of the Sey patient variant. Overall, we have shown successful encapsulation, transfection, and ex vivo genomic editing of an aniridia variant by ABE8e-RNP-LNPs in a clinically relevant cell type.
Discussion
We designed the CHuMMMs minimal-humanization strategy to answer the challenge of demonstrating efficacy of CRISPR-based therapies in animal in vitro, ex vivo, and in vivo model systems, while binding human DNA to facilitate rapid translation to the clinic. Here, we demonstrate the ease, functional tolerance, and usefulness of CHuMMMs, by developing a CRISPR-based therapeutic strategy for congenital aniridia. With ease we established three types of CHuMMMs cell lines, all homozygous for a humanized CRISPR “landing pad” of only 312 bp at exon 9 of Pax6 (He9). Furthermore, they were homozygous variant (He9−/He9−), heterozygous (He9−/He9+), and homozygous nonvariant (He9+/He9+), for the most common aniridia patient variant, c.718C > T. Functional tolerance was demonstrated by generating a CHuMMMs mouse strain [homozygous nonvariant (He9+/He9+)] that showed humanization alone did not disrupt Pax6 gene function in vivo. Finally, we used the cell lines to compare the efficacy of five different CRISPR enzymes. Overall, this work demonstrates the suitability of the CHuMMMs strategy, which can be applied widely to increase the value of all types of animal models for preclinical CRISPR therapy development for genetic diseases.
When comparing the efficacy of CRISPR enzymes in the He9 CHuMMMs ESCs we found that ABE8e is the optimal enzyme for gene editing at Pax6 exon 9 patient variant c.718C > T. ABE8e was the enzyme that demonstrated the highest average genomic correction of the patient variant at 76.8 ± 0.48%. ABE8e also showed the lowest editing of the nonvariant chromosome at 0.545 ± 0.73%. This differential in editing activity is ideal for the purposes of treating aniridia patient cells, as they are heterozygous for the pathogenic variant. By comparing the editing activity in the three types of He9 ESC lines, we found editing was additive, without interaction when both variant and nonvariant chromosomes were present. In addition to the variant correction, we observed bystander editing by ABE8e. Fortunately, the bystander edit results in a synonymous mutation, and therefore does not lead to an amino acid change. Overall, this work shows that ABE8e can correct the most commonly reported causal variant of aniridia. In addition, while other studies have shown successful genomic editing using ABE8e delivered in vitro as either plasmid, mRNA, or RNP [38, 57,58,59,60,61], this is the first study to successfully edit the DNA of mouse ESCs utilizing the safer RNP approach.
Once we determined that ABE8e was the optimal CRISPR-based enzyme for gene editing at this locus, we sought to investigate a translatable delivery system to a more clinically relevant cell type. In choosing between rAAV and LNPs, our data showing the importance of sgRNA modification strongly favored the use of LNPs. Our results show the first successful delivery and genomic editing by the CRISPR ABE8e encapsulated as an LNP-RNP. As a clinically relevant cell type we choose ex vivo mouse primary cortical neurons. This necessitated shifting to another patient variant, which strengthens the applicability of our work showing correction of two aniridia patient variants, but ABE8e was only able to correct this second genomic variant to an encoded missense mutation. Overall, correction dropped from 76.8% by electroporation in ESCs to correct the c.718C > T (p.R240X) genomic variant to wild type, versus 24.8% by LNP-RNPs in primary neurons to alter the c.580G > T (p.G194X) variant to Pax6 protein with a missense arginine. We hypothesize this difference may be due to a combination of transfection methodology, cell type, sgRNA sequence, DNA-sequence at the target site, and instability of the missense-carrying Pax6 protein.
There are two main limitations of this study to consider. First, there are over 600 PAX6 causal variants for aniridia [8]. Here we have demonstrated a high level of correction of the most common variant, but even that variant only represents one of four that together account for more than 20% of aniridia cases [7, 62]. We anticipated that developing a gene therapy for this variant will pave the way for similar personalized medicine approaches for other variants, but that needs future realization. Second, as with all animal model systems, it is not possible to test the off-target impact of CRISPR enzymes, which must be studied for the entire human genome. However, the finding that the base editor ABE8e was the optimal CRISPR-based enzyme is advantageous, since this enzyme does not create DSBs and is well known for minimal off-target effects [38].
There are two potential approaches for clinical application of our optimized CRISPR therapeutic strategy: ex vivo or in vivo. The first application may be ex vivo autologous cell therapy with correction of patient-derived cells from either the retina or cornea prior to transplantation into the aniridic eye [63]. A second application may be in vivo administration, to deliver the CRISPR therapy to multiple tissues of the patient eye [63]. Nonetheless, both administration approaches require additional study to demonstrate safety and efficacy for clinical translation.
Conclusions
The results of this study demonstrate support for the hypothesis that a CRISPR gene therapy can be developed and optimized in humanized mouse embryonic stem cells that will be able to distinguish between an aniridia patient variant and nonvariant chromosomes. We demonstrated the usefulness of the CHuMMMs approach, and showed the first genomic editing by ABE8e encapsulated as an LNP-RNP. Overall, this study demonstrates successful ABE8e-mediated editing of two aniridia patient variants, and thus lays the foundation for further preclinical in vivo mouse studies to rescue Pax6 expression and prevent disease phenotype.
References
Hingorani M, Hanson I, van Heyningen V. Aniridia. Eur J Hum Genet. 2012;20(10):1011–7.
Moosajee M, Hingorani M, Moore AT, et al. PAX6-related aniridia. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, et al., editors. Gene reviews. Seattle, WA: University of Washington; 1993.
Landsend ECS, Lagali N, Utheim TP. Congenital aniridia—a comprehensive review of clinical features and therapeutic approach. Surv Ophthalmol. 2021;66(6):1031–50.
Latta L, Figueiredo FC, Ashery-Padan R, Collinson JM, Daniels J, Ferrari S, et al. Pathophysiology of aniridia-associated keratopathy: developmental aspects and unanswered questions. Ocul Surf. 2021;22:245–56.
Schlotzer-Schrehardt U, Latta L, Giessl A, Zenkel M, Fries FN, Kasmann-Kellner B, et al. Dysfunction of the limbal epithelial stem cell niche in aniridia-associated keratopathy. Ocul Surf. 2021;21:160–73.
Heavner W, Pevny L. Eye development and retinogenesis. Cold Spring Harb Perspect Biol. 2012;4(12): a008391.
Lima Cunha D, Arno G, Corton M, Moosajee M. The spectrum of PAX6 mutations and genotype–phenotype correlations in the eye. Genes (Basel). 2019;10(12):1050.
Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–8.
Gregory-Evans CY, Wang X, Wasan KM, Zhao J, Metcalfe AL, Gregory-Evans K. Postnatal manipulation of Pax6 dosage reverses congenital tissue malformation defects. J Clin Invest. 2013;124(1):111–6.
Pedersen HR, Baraas RC, Landsend ECS, Utheim OA, Utheim TP, Gilson SJ, et al. PAX6 genotypic and retinal phenotypic characterization in congenital aniridia. Invest Ophthalmol Vis Sci. 2020;61(5):14.
Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT. LOVD v.2.0: the next generation in gene variant databases. Hum Mutat. 2011;32(5):557–63.
Guo R, Zhang X, Liu A, Ji J, Liu W. Novel clinical presentation and PAX6 mutation in families with congenital aniridia. Front Med (Lausanne). 2022;9:1042588.
Kit V, Lima Cunha D, Hagag AM, Moosajee M. Longitudinal genotype-phenotype analysis in 86 PAX6-related aniridia patients. JCI Insight. 2021. https://doi.org/10.1172/jci.insight.148406.
Tyner C, Barber GP, Casper J, Clawson H, Diekhans M, Eisenhart C, et al. The UCSC genome browser database: 2017 update. Nucleic Acids Res. 2017;45(D1):D626–34.
Cole JD, McHaney KM, Rabiee B, Gao J, Rodriguez C, Miller DA, et al. Long-term retinal protection by MEK inhibition in Pax6 haploinsufficiency mice. Exp Eye Res. 2022;218: 109012.
Daruich A, Duncan M, Robert MP, Lagali N, Semina EV, Aberdam D, et al. Congenital aniridia beyond black eyes: from phenotype and novel genetic mechanisms to innovative therapeutic approaches. Prog Retin Eye Res. 2022. https://doi.org/10.1016/j.preteyeres.2022.101133.
Abdolkarimi D, Cunha DL, Lahne M, Moosajee M. PAX6 disease models for aniridia. Indian J Ophthalmol. 2022;70(12):4119–29.
Wang X, Gregory-Evans K, Wasan KM, Sivak O, Shan X, Gregory-Evans CY. Efficacy of postnatal in vivo nonsense suppression therapy in a Pax6 mouse model of aniridia. Mol Ther Nucleic Acids. 2017;7:417–28.
Hill RE, Favor J, Hogan BL, Ton CC, Saunders GF, Hanson IM, et al. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354(6354):522–5.
Hickmott JW, Gunawardane U, Jensen K, Korecki AJ, Simpson EM. Epistasis between PAX6Sey and genetic background reinforces the value of defined hybrid mouse models for therapeutic trials. Gene Ther. 2018;25(8):524–37.
Mirjalili Mohanna SZ, Hickmott JW, Lam SL, Chiu NY, Lengyell TC, Tam BM, et al. Germline CRISPR/Cas9-mediated gene editing prevents vision loss in a novel mouse model of aniridia. Mol Ther Methods Clin Dev. 2020;17:478–90.
Tay LS, Palmer N, Panwala R, Chew WL, Mali P. Translating CRISPR-Cas therapeutics: approaches and challenges. CRISPR J. 2020;3(4):253–75.
Zhu F, Nair RR, Fisher EMC, Cunningham TJ. Humanising the mouse genome piece by piece. Nat Commun. 2019;10(1):1845.
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–21.
Maeder ML, Stefanidakis M, Wilson CJ, Baral R, Barrera LA, Bounoutas GS, et al. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat Med. 2019;25(2):229–33.
Li T, Yang Y, Qi H, Cui W, Zhang L, Fu X, et al. CRISPR/Cas9 therapeutics: progress and prospects. Signal Transduct Target Ther. 2023;8(1):36.
Yin S, Zhang M, Liu Y, Sun X, Guan Y, Chen X, et al. Engineering of efficiency enhanced Cas9 and base editors with improved gene therapy efficacies. Mol Ther. 2022;31(3):744–59.
Shin JW, Hong EP, Park SS, Choi DE, Seong IS, Whittaker MN, et al. Allele-specific silencing of the gain-of-function mutation in Huntington’s disease using CRISPR/Cas9. JCI Insight. 2022. https://doi.org/10.1172/jci.insight.141042.
Walton RT, Christie KA, Whittaker MN, Kleinstiver BP. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020;368(6488):290–6.
Cao X, Guo J, Huang S, Yu W, Li G, An L, et al. Engineering of near-PAMless adenine base editor with enhanced editing activity and reduced off-target. Mol Ther Nucleic Acids. 2022;28:732–42.
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–308.
Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 2017;551(7681):464–71.
Li H, Yang Y, Hong W, Huang M, Wu M, Zhao X. Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct Target Ther. 2020;5(1):1.
Jo DH, Jang HK, Cho CS, Han JH, Ryu G, Jung Y, et al. Visual function restoration in a mouse model of Leber congenital amaurosis via therapeutic base editing. Mol Ther Nucleic Acids. 2023;31:16–27.
Zhang Y, Nishiyama T, Li H, Huang J, Atmanli A, Sanchez-Ortiz E, et al. A consolidated AAV system for single-cut CRISPR correction of a common Duchenne muscular dystrophy mutation. Mol Ther Methods Clin Dev. 2021;22:122–32.
Xiao Q, Xu Z, Xue Y, Xu C, Han L, Liu Y, et al. Rescue of autosomal dominant hearing loss by in vivo delivery of mini dCas13X-derived RNA base editor. Sci Transl Med. 2022;14(654):eabn0449.
Dong JY, Fan PD, Frizzell RA. Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum Gene Ther. 1996;7(17):2101–12.
Richter MF, Zhao KT, Eton E, Lapinaite A, Newby GA, Thuronyi BW, et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat Biotechnol. 2020;38(7):883–91.
Chamberlain K, Riyad JM, Weber T. Expressing transgenes that exceed the packaging capacity of adeno-associated virus capsids. Hum Gene Ther Methods. 2016;27(1):1–12.
Koblan LW, Erdos MR, Wilson C, Cabral WA, Levy JM, Xiong ZM, et al. In vivo base editing rescues Hutchinson–Gilford progeria syndrome in mice. Nature. 2021;589(7843):608–14.
Finn JD, Smith AR, Patel MC, Shaw L, Youniss MR, van Heteren J, et al. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 2018;22(9):2227–35.
Gillmore JD, Gane E, Taubel J, Kao J, Fontana M, Maitland ML, et al. CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N Engl J Med. 2021;385(6):493–502.
Han JP, Kim M, Choi BS, Lee JH, Lee GS, Jeong M, et al. In vivo delivery of CRISPR-Cas9 using lipid nanoparticles enables antithrombin gene editing for sustainable hemophilia A and B therapy. Sci Adv. 2022;8(3):eabj6901.
Herrera-Barrera M, Ryals RC, Gautam M, Jozic A, Landry M, Korzun T, et al. Peptide-guided lipid nanoparticles deliver mRNA to the neural retina of rodents and nonhuman primates. Sci Adv. 2023;9(2):eadd4623.
Zhang D, Wang G, Yu X, Wei T, Farbiak L, Johnson LT, et al. Enhancing CRISPR/Cas gene editing through modulating cellular mechanical properties for cancer therapy. Nat Nanotechnol. 2022;17(7):777–87.
Jang HK, Jo DH, Lee SN, Cho CS, Jeong YK, Jung Y, et al. High-purity production and precise editing of DNA base editing ribonucleoproteins. Sci Adv. 2021;7(35):2661.
Chen Q, Zhang Y, Yin H. Recent advances in chemical modifications of guide RNA, mRNA and donor template for CRISPR-mediated genome editing. Adv Drug Deliv Rev. 2021;168:246–58.
Peeters SB, Korecki AJ, Simpson EM, Brown CJ. Human cis-acting elements regulating escape from X-chromosome inactivation function in mouse. Hum Mol Genet. 2018;27(7):1252–62.
Yang GS, Banks KG, Bonaguro RJ, Wilson G, Dreolini L, de Leeuw CN, et al. Next generation tools for high-throughput promoter and expression analysis employing single-copy knock-ins at the Hprt1 locus. Genomics. 2009;93:196–204.
Kluesner MG, Nedveck DA, Lahr WS, Garbe JR, Abrahante JE, Webber BR, et al. EditR: a method to quantify base editing from Sanger sequencing. CRISPR J. 2018;1(3):239–50.
Huang TP, Newby GA, Liu DR. Precision genome editing using cytosine and adenine base editors in mammalian cells. Nat Protoc. 2021;16(2):1089–128.
Mirjalili Mohanna SZM, Djaksigulova D, Hill AM, Wagner PK, Simpson EM, Leavitt BR. LNP-mediated delivery of CRISPR RNP for wide-spread in vivo genome editing in mouse cornea. J Control Release. 2022;350:401–13.
Bradley A, Evans M, Kaufman MH, Robertson E. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature. 1984;309(5965):255–6.
Mathew SM. Strategies for generation of mice via CRISPR/HDR-mediated knock-in. Mol Biol Rep. 2023;50(4):3189–204.
Gertsenstein M, Mianne J, Teboul L, Nutter LMJ. Targeted mutations in the mouse via embryonic stem cells. Methods Mol Biol. 2020;2066:59–82.
Vakulskas CA, Dever DP, Rettig GR, Turk R, Jacobi AM, Collingwood MA, et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat Med. 2018;24(8):1216–24.
Kulcsar PI, Talas A, Ligeti Z, Krausz SL, Welker E. SuperFi-Cas9 exhibits remarkable fidelity but severely reduced activity yet works effectively with ABE8e. Nat Commun. 2022;13(1):6858.
Alves CRR, Ha LL, Yaworski R, Lazzarotto CR, Christie KA, Reilly A, et al. Base editing as a genetic treatment for spinal muscular atrophy. biorXiv. 2023. https://doi.org/10.1101/2023.01.20.524978.
Newby GA, Yen JS, Woodard KJ, Mayuranathan T, Lazzarotto CR, Li Y, et al. Base editing of haematopoietic stem cells rescues sickle cell disease in mice. Nature. 2021;595(7866):295–302.
Haideri T, Howells A, Jiang Y, Yang J, Bao X, Lian XL. Robust genome editing via modRNA-based Cas9 or base editor in human pluripotent stem cells. Cell Rep Methods. 2022;2(9): 100290.
Sheriff A, Guri I, Zebrowska P, Llopis-Hernandez V, Brooks IR, Tekkela S, et al. ABE8e adenine base editor precisely and efficiently corrects a recurrent COL7A1 nonsense mutation. Sci Rep. 2022;12(1):19643.
Blanco-Kelly F, Tarilonte M, Villamar M, Damian A, Tamayo A, Moreno-Pelayo MA, et al. Genetics and epidemiology of aniridia: updated guidelines for genetic study. Arch Soc Esp Oftalmol (Engl Ed). 2021;96(Suppl 1):4–14.
Salman M, Verma A, Singh VK, Jaffet J, Chaurasia S, Sahel DK, et al. New frontier in the management of corneal dystrophies: basics, development, and challenges in corneal gene therapy and gene editing. Asia Pac J Ophthalmol (Phila). 2022;11(4):346–59.
Acknowledgements
Technical Assistance. We want to thank Dr. Nada Lallous and Mr. Joseph Lee at the Vancouver General Hospital Gobind Khorana Protein Engineering Core for expertise in purification of the ABE8e protein, Mr. Austin Hill at Incisive Genetics Inc. for contributing to contracting and funding acquisition, Dr. Sunita Sinha at the University of British Columbia Sequencing and Bioinformatics Consortium for Sanger sequencing, Ms. Marketa Hlavon and Mr. Cillein Thorne at the British Columbia Children’s Hospital Research Institute (BCCHRI) Sequencing and Bioanalyzer Core also for Sanger sequencing, and Ainsley Coquinco at Incisive Genetics Inc. for cortical neuron cultures.
Funding. This work was funded by a Canadian Institutes of Health Research (CIHR) grant (PJT-159484) awarded to E.M.S., Incisive Genetics Inc., Vancouver, British Columbia, Canada (AWD-021095), and a BCCHRI Brain, Behaviour & Development Trainee Boost (AWD-017347) awarded to B.A.A. The Phoenix Micron Anterior Segment Imaging system and SomnoSuite were funded by the Canada Foundation for Innovation (35143), British Columbia Knowledge Development Fund, and BCCHRI (CRG74760). The Rapid Service Fee was funded by CIHR (PJT-159484) and Incisive Genetics Inc.
Author Contributions. Conceptualization, Elizabeth M. Simpson.; Data curation, Bethany A. Adair and Andrea J. Korecki; Formal analysis, Bethany A. Adair; Funding acquisition, Bethany A. Adair, Blair R. Leavitt, and Elizabeth M. Simpson; Investigation and Methodology, Bethany A. Adair, Andrea J. Korecki, Diana Djaksigulova, Pamela K. Wagner, Nina Y. Chiu, Siu Ling Lam, and Tess C. Lengyell; Supervision, Blair R. Leavitt, and Elizabeth M. Simpson; Visualization, Bethany A. Adair, Writing – original draft, Bethany A. Adair; Writing – review & editing, Andrea J. Korecki, Tess C. Lengyell, and Elizabeth M. Simpson.
Disclosures. Andrea J. Korecki, Diana Djaksigulova, Nina Y. Chiu, Siu Ling Lam, and Tess C. Lengyell, declare no competing interests. Bethany A. Adair, Pamela K. Wagner, Blair R. Leavitt, Elizabeth M. Simpson, and the University of British Columbia (UBC) have filed patent application (PCT/US2019/0055472 and PCT/IB2023/051742) on the delivery of ribonucleoprotein complexes in lipid nanoparticles. Pamela K. Wagner, and Blair R. Leavitt and are co-founders of Incisive Genetics Inc., which has the exclusive license to the intellectual property from UBC.
Compliance with Ethics Guidelines. All animals were housed and bred in the pathogen-free Transgenic Animal facility at the Centre for Molecular Medicine and Therapeutics (CMMT) of the University of British Columbia (UBC). All mouse work was performed following protocols approved by the UBC Animal Care Committee (protocol #s A21-0410, A21-0184), in accordance with guidelines determined by the Canadian Council on Animal Care.
Data Availability. Authors can confirm that all relevant data are included in the article and/or its supplementary material files or from the corresponding author, by request.
Author information
Authors and Affiliations
Corresponding author
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Adair, B.A., Korecki, A.J., Djaksigulova, D. et al. ABE8e Corrects Pax6-Aniridic Variant in Humanized Mouse ESCs and via LNPs in Ex Vivo Cortical Neurons. Ophthalmol Ther 12, 2049–2068 (2023). https://doi.org/10.1007/s40123-023-00729-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40123-023-00729-6