Abstract
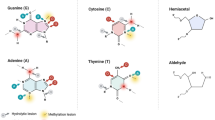
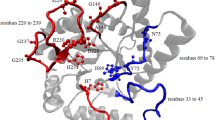
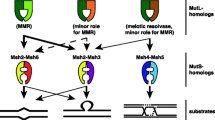
DNA repair refers to a collection of processes by which a cell identifies and corrects damage to DNA molecules that encodes its genome. In this study, we examined evolution of O6-Methylguanine DNA alkyltransferase (MGMT), Xeroderma pigmentosum group D (XPD) protein, G/T mismatch-specific DNA glycosylases, MutS DNA repair proteins of Escherichia coli, Pyrococcus kodakaraensis, Saccharomyces cerevisae, Drosophila melanogester, Mus musculus, and Homo sapiens, which are involved in direct repair, Nucleotide excision repair, base excision repair, and mismatch repair, respectively. Sequence and domain analysis of these proteins indicates that during the course of evolution catalytic residues in the catalytic domain remain conserved. Phylogenetic tree analysis suggested that MGMT proteins of human and mouse have archaeal origin, whereas XPD proteins which are responsible for nucleotide excision repair evolved progressively from lower organism to higher organism. G/T mismatch-specific DNA glycosylases belong to family 1, family 2, and family 4 of uracil-DNA glycosylases superfamily. Family 2 proteins have broader substrate specificity in comparison to proteins of other families. In eukaryotic organisms, prokaryotic MUTS genes are duplicated and various paralogs are present. No unified evolution mechanism can explain the evolution of all these DNA repair proteins. Large sequence variation is observed among same DNA repair proteins of different organisms. However, residues involved DNA repair work and DNA binding remain conserved during the course of evolution.





Similar content being viewed by others
References
Allen DJ, Makhov A, Grilley M, Taylor J, Thresher R, Modrich P, Griffith JD (1997) MutS mediates heteroduplex loop formation by a translocation mechanism. The Embo J 16:4467–4476
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399
Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N (2010) ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucl Acids Res 38(Web Server issue):W529–W533
Bai H, Lu AL (2007) Physical and functional interactions between Escherichia coli MutY glycosylase and mismatch repair protein MutS. J Bacteriol 189(3):902–910
Bertrand P, Tishkoff DX, Filosi N, Dasgupta R, Kolodner RD (1998) Physical interaction between components of DNA mismatch repair and nucleotide excision repair. Proc Natl Acad Sci USA 95:14278–14283
Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the clustal series of programs. Nucl Acids Res 31:3497–3500
Dexheimer TS (2013) DNA Repair Pathways and Mechanisms. In: Mathews LA, Cabarcas SM, Hurt EM (eds) DNA repair of cancer stem cells. Springer, Netherlands, pp 19–32
Dmitry GV, Kosuke M (1997) DNA repair enzymes. Curr Opi Struc Bio 7:103–109
Dufner P, Marra G, Schle MR, Jiricny J (2000) Mismatch recognition and DNA-dependent stimulation of the ATPase activity of hMutSa is abolished by a single mutation in the hMSH6 subunit. J Bio Chem 275(47):36550–36555
Eisen JA (1998) A phylogenomic study of the MutS family of proteins. Nucl Acids Res 26:4291–4300
Fang Q, Kanugula S, Pegg AE (2005) Function of Domains of Human O6-Alkylguanine-DNA Alkyltransferase. Biochemistry 44:15396–15405
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Finn RD, Tate J, Mistry J, Coggill PC, Sammut JS, Hotz HR, Ceric GKF, Eddy SR, Sonnhammer EL, Bateman A (2008) The Pfam protein families database. Nucl Acids Res 36(Database Issue):D281–D288
Fishel R, Wilson T (1997) MutS homologs in mammalian cells. Curr Opin Gen Dev 7(1):105–113
Friedberg EC (2003) DNA damage and repair. Nature 421:436–440
Gellon L, Werner M, Boiteux S (2002) Ntg2p, a Saccharomyces cerevisiae DNA N-glycosylase/apurinic or apyrimidinic lyase involved in base excision repair of oxidative DNA damage, interacts with the DNA mismatch repair protein Mlh1p. Identification of a Mlh1p binding motif. J Biol Chem 277:29963–29972
Hardeland U, Bentele M, Lettieri T, Steinacher R, Jiricny J, Schär P (2001) Thymine DNA glycosylase. Prog Nucl Acid Res Mol Biol 68:235–253
Irigaray P, Belpomme D (2010) Basic properties and molecular mechanisms of exogenous chemical carcinogens. Carcinogenesis 31(2):135–148
Jane EAW, Anthony EP, Peter CEM (2000) Crystal structure of the human O6-alkylguanine-DNA alkyltransferase. Nucl Acids Res 28(2):393–401
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein Sequences. Comput Appl Biosc 8:275–282
Jorg S, Frank M, Peer B, Chris PP (1998) SMART, a simple modular architecture research tool: Identification of signaling domains. Proc Natl Acad Sci USA 95:5857–5864
Kolodner RD (1995) Mismatch repair: mechanisms and relationship to cancer susceptibility. Trends Biochem Sci 20:397–401
Kolodner R (1996) Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev 10:1433–1442
Kovtun IV, McMurray CT (2007) Crosstalk of DNA glycosylases with pathways other than base excision repair. DNA Repair 6:517–529
Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, De Wind N, Sixma TK (2000) The crystal structure of DNA mismatch repair protein MutS binding to a G × T mismatch. Nature 407(6805):711–717
Lindahl T, Wood RD (1999) Quality control by DNA repair. Science 286:1897–1905
Madeleine HM, Jacqueline M, Gulb EJD, Bruce D, Peter MCE (1994) Crystal structure of a suicidal DNA repair protein: the Ada 06-methylguanine-DNA methyltransferase from Eco1i. The Embo J 13(7):1495–1501
Marchler-Bauer A, Bryant SH (2004) CD Search: protein domain annotations on the fly. Nucl Acids Res 32(Web Server issue):W327–W331
Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, Ke Z, Krylov D, Lanczycki CJ, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Thanki N, Yamashita RA, Yin JJ, Zhang D, Bryant SH (2007) CDD: a conserved domain database for interactive domain family analysis. Nucl Acids Res 35(Database Issue):D237–D240
Mazon G, Philippin G, Cadet J, Gasparutto D, Fuchs RP (2009) The alkyltransferase-like ybaZ gene product enhances nucleotide excision repair of O6-alkylguanine adducts in E. coli. DNA Repair 8:697–703
McCulloch SD, Kunkel TA (2008) The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res 18(1):148–161
Millar CB, Guy J, Sansom OJ, Selfridge J, MacDougall E, Hendrich B, Keightley PD, Bishop SM, Clarke AR, Bird A (2002) Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science 297:403–405
Panier S, Boulton SJ (2014) Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol 15(1):7–18
Pearl LH (2000) Structure and function in the uracil-DNA glycosylase superfamily. Mutat Res 460:165–181
Pegg AE (2000) Repair of O(6)-alkylguanine by alkyltransferases. Mutat Res 462:83–100
Rada C, Di Noia JM, Neuberger MS (2004) Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol Cell 16:163–171
Rasimas JJ, Kanugula S, Dalessio PM, Ropson IJ, Pegg AE (2003) Effects of zinc occupancy on human O6-alkylguanine-DNA alkyltransferase. Biochemistry 42:980–990
Sancar A (1995) DNA repair in humans. Annu Rev Genet 29:69–105
Saparbaev M, Laval J (1998) 3, N 4-ethenocytosine, a highly mutagenic adduct, is a primary substrate for Escherichia coli double-stranded uracil-DNA glycosylase and human mismatch-specific thymine-DNA glycosylase. Proc Natl Acad Sci USA 95:8508–8513
Shimizu Y, Iwai S, Hanaoka F, Sugasawa K (2003) Xeroderma pigmentosum group C protein interacts physically and functionally with thymine DNA glycosylase. EMBO J 22:164–173
Singleton MR, Dillingham MS, Wigley DB (2007) Structure and mechanism of helicases and nucleic acid tanslocases. Annu Rev Biochem 76:23–50
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tornaletti S, Maeda LS, Lloyd DR, Reines D, Hanawalt PC (2001) Effect of thymine glycol on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II. J Biol Chem 276:45367–45371
Tubbs JL, Tainer JA (2010) Alkyltransferase-like proteins: Molecular switches between DNA repair pathways. Cell Mol Life Sci 67(22):3749–3762
Tubbs JL, Latypov V, Kanugula S, Butt A, Melikishvili M, Kraehenbuehl R, Fleck O, Marriott A, Watson AJ, Verbeek B, McGown G, Thorncroft M, Santibanez-Koref MF, Millington C, Arvai AS, Kroeger MD, Peterson LA, Williams DM, Fried MG, Margison GP, Pegg AE, Tainer JA (2009) Flipping of alkylated DNA damage bridges base and nucleotide excision repair. Nature 459:808–813
Vincent WF, Neale PJ (2000) Mechanisms of UV damage to aquatic organisms. In: Mora SD, Demers S, Vernet M (eds) The effects of UV radiation on marine ecosystems. Cambridge University Press, Cambridge, pp 149–176
Viswanathan A, Doetsch PW (1998) Effects of nonbulky DNA base damages on Escherichia coli RNA polymerase-mediated elongation and promoter clearance. J Biol Chem 273:21276–21281
Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Paul LM, Beese LS (2007) Structure of the human MutS alpha DNA lesion recognition complex. Mol Cell 26:579–592
Winkler GS, Araujo SJ, Fiedler U, Vermulen W, Coin F (2000) TFIIH with inactive Xpd helicase functions in transcription initiation but it is defective in DNA repair. J Biol Chem 275:4258–4266
Wolski SC, Kuper J, Nzelmann PH, Truglio JJ, Croteau DL, Houten BV, Kisker C (2008) Crystal structure of the FeS cluster-containing nucleotide excision repair helicase XPD. Plos Bio 6(6):e149
Acknowledgments
Swarna Kanchan acknowledges the support in the form of fellowship given by University Grant Commission, New Delhi, Govt. of India for Basic Science Research (BSR) fellowship. Authors would also like to acknowledge the infrastructural facilities provided by Department of Biological Sciences, Birla Institute of Technology and Science, Pilani, Rajasthan, India.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kanchan, S., Mehrotra, R. & Chowdhury, S. Evolutionary pattern of four representative DNA repair proteins across six model organisms: an in silico analysis. Netw Model Anal Health Inform Bioinforma 3, 70 (2014). https://doi.org/10.1007/s13721-014-0070-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13721-014-0070-1