Abstract
Honey bees are very important eusocial insects and are involved in the pollination of many plants. Queen bees and worker bees can develop from the same fertilized eggs and are thus genetically identical despite their substantial behavioral and physiological differences. The mechanism governing developmental differences between worker and queen bees has always attracted much interest. While there are several reports on messenger RNA (mRNA) expression related to caste differentiation or microRNA (miRNA) expression in one time point of caste differentiation, no systematic investigation of the dynamic expression of small RNAs along with these two caste development has, thus far, been carried out. In this study, we present the dynamic expression profiles of queen and worker bee small RNAs and show caste-specific miRNA expression patterns between them, indicating that miRNAs may be related to the differential development of worker and queen bee larvae. Results presented here will make a valuable contribution to understanding of the caste switch between worker and queen bees.
Similar content being viewed by others

1 Introduction
The honeybee is a eusocial insect that exhibits complete metamorphosis. In a bee colony, there are normally three social classes: drones, workers, and a single queen. The drones are haploid, short-lived male bees that develop from unfertilized eggs and normally make up 500–2000 individuals in a normal hive. Worker and queen bees both develop from fertilized eggs and are thus genetically identical despite their substantial behavioral and physiological differences. Worker bees are sterile, diploid females, which make up the bulk of the hive (40–100 thousand individuals). They have underdeveloped reproductive organs but have keen sensory apparatus and a strong ability to act as guards for the colony. The queen, on the other hand, is larger in size with a longer life span and has an extremely high reproductive capacity (Evans and Wheeler 1999; Hartfelder et al. 2015; Kamakura 2011; Leimar et al. 2012; Page and Peng 2001; Shuel and Dixon 1960). Although the proportion of queen bees in a colony is very small, the generation of queens is not random. The differentiation of queen bees is controlled by nurse bees (Moritz et al. 2005). After the third day of larval development, differences in nutritional or other environmental factors cause the development of workers and queen larvae to take distinct developmental paths (Dedej et al. 1998; Evans and Wheeler 2001; Guo et al. 2013; Kucharski et al. 2008). Artificial queens rearing with worker larvae grafted from the fourth day or days later would develop significantly decreased number of ovarioles and basitarsal index (Dedej et al. 1998) when compared to ones rearing from early stages, showing that the age around 3.5 days is committed to the development of honey bee as either worker or queen. The expression profiling of bee larvae collected at six 1-day intervals from the embryo stage to the middle of the fifth and final instar suggested most gene-expression changes associated with caste programs occurred at day 3 or later (Evans and Wheeler 2001), although the expressional differences of genes between worker and queen-destined larvae may happen at 6 h after hatching (Cameron et al. 2013).
The mechanism governing the developmental difference between worker and queen bees has therefore attracted much interest. It has been shown that ecdysone, juvenile hormone (JH), TOR pathway, epigenetic modification, royalactin, and insulin are all involved in caste determination in insects (Capella and Hartfelder 1998; Hartfelder et al. 2015; Kamakura 2011; Kucharski et al. 2008; Leimar et al. 2012; Patel et al. 2007; Pinto et al. 2002; Wheeler et al. 2006). During the third day, high levels of JH induce the development of the queen caste, whereas low levels of JH result in the development of other worker castes. In addition, the sugar content in the nutrition provided to the developing larvae affects the rate of food intake in honey bee larvae, which in turn affect corpora allata activity and switch in caste determination. With gene functional experiments, several groups reported the roles of the single gene on the caste differentiation of two bee sisters (Kamakura 2011; Kucharski et al. 2008; Patel et al. 2007). When TOR pathway for queen-destined larvae was blocked by TOR inhibitor or siRNA, these queen-destined larvae would develop worker characters and even emerge with fully developed worker morphology (Patel et al. 2007). Royalactin was reported to induce queen differentiation as well (Kamakura 2011) although others have debated this (Leimar et al. 2012). In contrast to the queen inducing genes, gene Dnmt3 has been shown to have a repressing role in the process of queen differentiation, and the majority of bee larvae with DNA methyltransferase Dnmt3 know-down emerged as queens with fully developed ovaries (Kucharski et al. 2008).
MicroRNAs (miRNAs) are approximately 22 nucleotides (nt) long RNA molecules which regulate the activity of messenger RNAs (mRNAs) by specific interactions with mRNA sequence elements (Bartel 2004; Farh et al. 2005). Mature miRNAs are processed by RNase Dicer, an RNase III endonuclease, from 70- to 90-nt long precursors (pre-miRNAs), and regulate mRNA stability and translational activity by guiding the RISC RNA-protein complex to its target mRNAs through complementary binding to specific sites on the 3′-UTR. Female mouse Dicer knock-out mutants are infertile (Escalier 2008; Hong et al. 2008; Luense et al. 2009). In Drosophila, knock-out mutants of the gene cluster comprising the miR-100, let-7, and miR-125 genes display reduced fertility, motility, and flight (Sokol et al. 2008). The microRNA bantam regulates cell growth and proliferation, organ and body size, and germ cell fate (Brennecke et al. 2003; Hipfner et al. 2002; Moberg and Hariharan 2003; Yang et al. 2009). The miR-210 is involved in the production and development of Drosophila ovaries and female gametes (Grun et al. 2005), and miR-9a strongly influences wing development (Biryukova et al. 2009).
Several studies about miRNA in honey bee have been reported recently. Chen et al. surveyed the miRNA expression of mixed RNAs from different development stages including egg, larva, pupa, and adult by using high-throughput system SOLiD, which opened the field of miRNA profiling in bees (Chen et al. 2010). Later on, Greenberg et al. and Liu et al. expression profiled the miRNAs for nurse and forager bees by using high-throughput system Illumina/Solexa in parallel and found that, while not a large quantity, a number of miRNAs were differentially expressed between these two types of worker bees with different division of labor (Greenberg et al. 2012; Liu et al. 2012). Zondag et al. found that 28 % of known miRNAs could be expressed in the early embryo (24–30 h), and these miRNAs played roles in patterning segmentation (Zondag et al. 2012). Shi et al. and Guo et al. demonstrated that miRNAs were present in bee jellies and these jelly miRNAs may play important functions in the caste development of bees (Guo et al. 2013; Shi et al. 2012). In addition, Shi et al. compared the miRNA profiles between queen larvae and worker larvae at 4 days old (Apis mellifera) and found that 37 miRNAs were differentially expressed between these two castes in this specific stage (Shi et al. 2015). However, until now, no systematic investigation of small RNAs has been carried out regarding the dynamic changes of small RNAs along with the caste differentiation. Here, we describe high-throughput sequencing of small RNAs expressed in the honeybee and the substantial differences in the expression profiles of such transcripts, which were found between developing worker and queen bee larvae.
2 Material and methods
2.1 Preparation of honeybee colonies and larval sample collection
Three apparently healthy 10-frame colonies of “Zhenongda No.1”—a high-yielding royal jelly breed of Apis mellifera ligustica (S. et al. 2002)— were maintained at the Huajiachi campus of Zhejiang University. In each colony, the queen laid eggs over a period of 24 h in one empty frame which was subsequently moved to an egg-free super-chamber. After 66 h (less than 18 hours after hatching), we transferred 150 larvae into queen cups to rear queens in each colony and put the queen cup frames into their corresponding colonies. The 40–60 worker larvae and queen larvae were collected from each colony after 4 days (73∼90 h after hatching), 5 days (97∼114 h after hatching), and 6 days (121∼138 h after hatching). The larval samples were collected into 50-mL tubes, immediately frozen in liquid nitrogen, and stored at −80 °C until being used for RNA extraction.
2.2 Library construction and sequencing
Total RNA was isolated from larvae of queen and worker bees on day 4, day 5, and day 6 using TRIzol, respectively. To construct the small RNA library, equal amounts of total RNA on day 4, day 5, and day 6 from queen and worker bees were pooled together respectively and were first size fractionated on a 15 % Tris-Borate-EDTA (TBE) urea polyacrylamide gel, and the 18–30-nt fraction was excised. The gel slice was incubated in 0.3 M NaCl with rotation at room temperature for 4 h to elute the small RNAs. The eluted RNAs were precipitated and washed using ethanol, and resuspended in ultrapure water. Gel-purified small RNA molecules were then ligated to 5′ RNA adapters using T4 RNA ligase, and the ligation products were gel fractionated and purified. 5′ RNA adapter-ligated small RNAs were ligated to 3′ RNA adapters. The final ligation products were purified, reverse-transcribed, and amplified by PCR. Amplified products were gel fractionated, purified on a 6 % polyacrylamide gel in TBE buffer, and then eluted in 0.3 M NaCl with rotation at room temperature for 4 h. The purified PCR products were then precipitated using ethanol and resuspended in nuclease-free water. These purified PCR products were then subjected to Illumina 1G Genome Analyzer. The small RNA expression profiling data used for this study have been deposited in the Gene Expression Omnibus (GEO) and are publically accessible through GEO (GSE52630).
2.3 Mapping small RNA reads
Before mapping small RNA reads, the low-quality read (containing any number of base N or more than 4 bases with quality score lower than 10 or more than 6 bases with quality score lower than 13) and 5′/3′ adaptor sequences were filtered and discarded. The remaining reads were then mapped to the honeybee genome (Consortium 2006) with SOAP (Li et al. 2008). Only perfect matches were accepted, and these were stored and used in the following analyses.
2.4 Recovery of known miRNAs by deep sequencing
To recover the known miRNAs in miRBase13.0, we downloaded the annotations of mature miRNAs and miRNA precursors, and aligned all the small RNA reads to miRNAs/hairpins from miRBase13.0 using BLAST with an E-value threshold of 0.01. Since one miRNA can be present in different isoforms, we used the sum of the expression levels of all isoforms of a given miRNA gene as a digital measure of its expression level.
2.5 Annotation and classification of small RNAs into different categories
In order to annotate the small RNAs against known noncoding RNAs, we collected all noncoding RNAs (ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), small nucleolar RNAs (snoRNAs), and small nuclear RNAs (snRNAs)) from Rfam and NCBI. To classify these small RNA reads, we aligned them to the above noncoding RNAs (ncRNA) groups using BLAST with an E-value threshold of 0.01 and no more than two mismatches, and then classified them into different categories according to the type of noncoding annotation they matched to. After annotating these small RNAs, we collected all the remaining unannotated small RNAs and used MIREAP to predict their likelihood of being miRNA genes based on canonical hairpin structure and energy. Since a given miRNA is present in different isoforms, we took the predominant miRNA isoform as the novel mature miRNA and calculated its expression level as the sum of the expression levels of all miRNA isoforms for that miRNA.
To explore the location relationship between the refGenes and known/novel miRNAs, we downloaded the honeybee refGene dataset from UCSC (Kuhn et al. 2009) and compared the genomic distribution between miRNAs and refGene loci.
2.6 Comparison of small RNA expression levels between worker and queen larvae
To explore the mechanism governing the differentiation of worker larvae and queen larvae, we need to identify the differences between worker and queen small RNA profiles. The Audic and Claverie test was used to determine the statistical significance of differences in gene expression for an individual gene between the two larval libraries (Audic and Claverie 1997).
2.7 Prediction of miRNA targets
In animals, miRNAs can bind to the 3′-UTRs of mRNAs, form miRNA-mRNA duplexes and block the functions of the mRNAs. We predicted the miRNA targets based on the 3′-UTR sequences of bee genes using miRanda (Betel et al. 2008).
2.8 Measurement of the expression levels of miRNAs by quantitative real-time PCR
To measure the expression levels of miRNAs across the different stages between two types of bee larvae, we started with 2.5-μg total RNA for each sample (fourth day queens, fifth day queens, sixth day queens, fourth day workers, fifth day workers, and sixth day workers), respectively. Reverse transcription and quantitative real-time PCR was then performed using NCode™ First-Strand cDNA Synthesis and qRT-PCR Kits (Invitrogen) according to the manufacturer’s protocol with duplicates (Online Resource ESM 1). To analyze the miRNA expression levels at different developmental stages (day 4, day 5, day 6) in workers and queens, the ΔCT values for different developmental stages were calibrated against the respective CT value of the day 4 sample in queens. qPCR results are presented as the fold expression relative to the day 4 sample in queens as performed in other study (Guo et al. 2013) unless otherwise stated.
3 Results
3.1 High-throughput sequencing of small RNAs in queen and worker bee
We used the Illumina/Solexa high-throughput platform (HTP) to sequence small RNAs from fourth to sixth day worker and queen bee larvae. After filtering out short- (<18 nt) and low-quality reads, 7,487,282 and 6,820,812 small RNA reads were left for worker and queen bee larvae, respectively, which were used for further analysis (Table I).
The length distribution of the small RNAs for both worker and queen larvae peaked at 22 nt, as is typical for animal miRNAs. The second peak at 16 nt was also evident in worker larvae. The number of small RNAs of size 18–21 nt was greater in queen larvae than in worker larvae, while numbers of small RNAs of other lengths were greater in worker larvae than in queen larvae (Figure 1).
Approximately ∼88 % of the small RNA reads corresponded to annotated miRNAs, around ∼2 % were tRNAs, ∼1.7 % were rRNA fragments, ∼0.4 % were snRNAs, and ∼0.04 % were snoRNAs. The remaining ∼12.4 % of the sequence reads did not correspond to any annotated transcribed sequences (Figure 2a, b, Table I).
Component of small RNA libraries (a, b) and genomic location of miRNAs (c, d) in worker and queen bee larvae. The distance within 2000 bases to transcription start site and end site of genes were defined as “upstream” and “downstream”; the distance longer than 2000 bases was designed as “intergenic” in c, d.
3.2 Honeybee miRNAs were mainly derived from gene introns and intergenic regions
The genomic positions and transcriptional mode of miRNA loci are variable. MicroRNA loci are found in intergenic regions, introns of coding genes, and in rare cases also in overlapping coding exons (Li et al. 2007; Rodriguez et al. 2004). Genomic analysis of the honey bee miRNAs showed that ∼40 % of bee miRNAs located in intronic regions of protein coding genes and ∼40 % of bee miRNAs located in intergenic regions of protein coding genes (Figure 2c, d).
3.3 Intronic miRNA loci may be independently transcribed
Whereas intergenic miRNAs are generally derived from independent precursors, intronic miRNAs may either be spliced out from the host gene pre-mRNA transcript or be processed from an independently transcribed precursor (Ying and Lin 2006). Comparison of the expression profiles of intronic miRNAs and their host genes (unpublished data) showed that, in general, less than 50 % of the intronic miRNAs were co-expressed (i.e., detected simultaneously) with their host genes. The expression levels of intronic miRNAs were also poorly correlated (r = −0.01) with their host genes, possibly suggesting that the expression of intronic miRNAs is generally not dependent on host gene expression in honey bees.
3.4 Expression of known and novel miRNAs
Fifty-eight honeybee miRNA genes are annotated in miRBase version 13.0. HTP sequencing of fourth- to sixth-day queen and worker bee larvae small RNAs yielded transcripts corresponding to 53 miRNA loci (Figure 3a). The expression levels of these 53 miRNA loci varied considerably between queen and worker bee larvae (Table II).
The miRBase v.13.0 does not annotate bee complementary miRNAs (also known as miRNA*s which originate from opposite arms of the same stem-loop hairpin sequences as known miRNAs and whose sequences are complementary to known miRNAs); however, of the 53 known miRNAs, we discovered 52 miRNA*s complementary to these known miRNAs. In addition, we also found substantial variation in miRNA* expression levels between the two small RNA libraries, and in nine cases, the miRNA* expression levels were even higher than those of the corresponding miRNAs (Online Resource ESM 2).
In order to identify possible novel miRNAs among the unannotated sequence reads, we filtered out all small RNAs that were against known ncRNAs. Around 71,050 and 37,622 newly identified small RNAs for worker and queen bee larvae, respectively, were found in this study (Table I), and these newly identified small RNAs were then subjected to miRNA prediction. To predict novel miRNA genes, we analyzed the flanking sequences of all the above small RNA loci for pre-miRNA hairpin structures. This resulted in the identification of 639 mature miRNA candidates (representing 585 miRNA precursor genes), of which 431 and 321 were present in the worker and queen bee larvae, respectively (Online Resource ESM 3). The comparison of expression levels of known and novel miRNAs showed that the expression of novel miRNA candidates was much lower than that of the known miRNAs.
In addition, these novel miRNAs showed strong differences in expression profiles between worker and queen bee larvae, and only 113 transcripts were present both in queen and in the worker bee larvae (Figure 3b). The remaining 526 miRNA candidates were only expressed in the worker (318) or in the queen (208) bee larvae, respectively, suggesting a possible relationship between miRNAs and the differential development of worker and queen bee larvae (Figure 3b).
Among the 639 miRNA candidates, there were 54 miRNA/miRNA pairs (Online Resource ESM 4 and ESM 5). This observation suggests that they may be processed from the same precursor transcripts via the miRNA pathway. The fact that their precursor sequences would allow formation of canonical pre-miRNA hairpin structures further validates the results. In addition, 28 of the 585 novel miRNA gene candidates analyzed by mirAlign were conserved in other organisms (Wang et al. 2005); 202 of the 585 miRNA candidates were predicted to be “real” miRNAs using miPred analysis (Online Resource ESM 3) (Jiang et al. 2007).
3.5 Worker and queen bee larvae differ in miRNA expression profiles
Worker and queen bee larvae showed substantial differences in miRNA expression levels, for example, the expression level of ame-let-7 in queen larvae was twice that of worker bee larvae, whereas the expression of ame-miR-263 in queen larva was only one fifth of that in worker bee larvae (Table II). By applying the Audic and Claverie test (Audic and Claverie 1997), we found that 23 known miRNAs were expressed at significantly higher levels (P < 0.05) in worker than in queen bee larvae and, by contrast, another 15 miRNAs were expressed at significantly higher levels in queen bee larvae than in worker bee larvae (Table II). Weaver et al. had surveyed the miRNA expression in a limited scale on 10 miRNAs identified by computational methods based on sequence homology and stem-loop scanning and successfully showed that several of these miRNAs had differential expression between queens and workers in the abdomen, head, pupa, and thorax (Weaver et al. 2007). Ame-miR-71 was found to have far stronger expression in abdomen of developing (pupal) workers than queens, while this miRNA was significantly highly expressed in worker larvae than in queen larvae by our small RNA-sequencing as well (P = 0). In contrast, ame-miR-iab-4 was stronger in pupa of queens than ones of workers, while this miRNAs was significantly higher in queen larvae than in the worker larvae (P = 3.30E-82) (Weaver et al. 2007).
JH has been shown to play an important role in the differential development of worker and queen bees (Rembold et al. 1974). Dozens of JH-related genes, which involved in JH signaling, biosynthesis, and metabolism, have been recently identified in the honey bee genome (Bomtorin et al. 2014; Cheng et al. 2014). One of them is juvenile hormone acid methyltransferase (NCBI Reference Sequence database: XM_001119986), which is differentially expressed during the caste differentiation (Li et al. 2013). A putative AMP-binding enzyme gene LOC726040 (XM_001121814) was detected to be JH responsive and also differentially expressed between queen and worker larvae (Barchuk et al. 2007). IAP2 (inhibitor of apoptosis 2, XM_396819), which can block cell death in fruit fly Drosophila melanogaster (Hay et al. 1995), may be able to response to JH as well, because JH affected the apoptosis of queen and worker ovaries during caste-specific differentiation (Capella and Hartfelder 1998). Given that genes related/responsive to JH levels are extremely important for the differential development of queen and worker bee larvae, we investigated miRNAs that might potentially target these three genes. All the seven miRNAs targeting the three genes mentioned above except for ame-miR-12 (Online Resource ESM 6) were expressed at low levels in queen bee larvae in comparison to worker bee larvae. This finding is in accordance with our hypothesis that the expression levels of JH-related/responsive genes and the miRNAs that potentially target them may show an opposite changing pattern, suggesting an actual regulatory relationship between these two sets of genes. In addition, there were numerous examples of miRNAs that showed substantial variation in expression levels between queen and worker bee larvae.
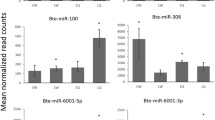
By small RNA sequencing of the mixture of equal amount of RNAs on day 4, day 5, and day 6 from queen and worker bees as the first step, we determined and screened the miRNAs expressed in queen and worker bees at three larval stages and approximately compared the expression differences between these two castes. In order to further explore the dynamic expression variations along with two caste development for queen and worker bees, we subsequently used quantitative RT-PCR (qPCR) to validate the expression of 23 known and 52 novel miRNAs in the fourth, fifth, and sixth day stages in queen and worker bee larvae along with the development time points. The qPCR results showed that there was considerable variation in miRNA expression both across stages and larval types, respectively. For instance, the expression level of ame-miR-1 in the fourth day worker bee larvae was five times more than that of day 6 larvae, whereas in queen bee larvae, the expression level of the same miRNA at the sixth day was seven times more than that it the fourth day. Comparison of the two larval types showed that the expression level of ame-miR-1 in the fourth day in worker bee larvae was 15 times that of the same day in queen bee larvae (Figure 4a). Moreover, the three miRNAs ame-miR-184, ame-miR-283, and ame-miR-10 showed a gradual increase in expression during queen bee larva development but decreased over the same period in worker bee larvae (Figure 4b, c). An opposite expression pattern of miRNAs between queen and worker bee was also demonstrated in ame-miR-279, ame-miR-87, and ame-miR-276 (Online Resource ESM 7). In addition, there were some other more complicated expression patterns along with the caste development of bee larvae. Twelve miRNAs including ame-bantam, ame-miR-100, ame-miR-12, ame-miR-71, ame-miR-263, ame-miR-277, ame-miR-14, ame-miR-190, ame-miR-8, ame-miR-275, ame-miR-375, and ame-miR-281 were shown to have a gradual decrease in expression during worker bee development but were shown to have an increased expression from day 4 to day 5 followed by a decrease from day 5 to day 6. For miRNA ame-miR-124 and ame-miR-305, the expression levels were slightly higher on day 5 than on day 4 and reached peak on day 5, followed by a decrease from day 5 to day 6 during worker bee development; the expression of these two miRNAs increased from day 4 to day 5 but kept similar level or was slightly dropped from day 5 to day 6 in queen bee (Online Resource ESM 7). Additional examples of distinct miRNA expression profiles could be found in Online Resource ESM 7 and ESM 8.
For the JH-related miRNAs, we performed qPCR to detect expression profiles of four of them in day 4, day 5, and day 6 worker and queen larvae (Online Resource ESM 9). All of these four miRNA genes reached the highest expression levels in day 4 worker bee larvae, and their expression gradually decreased from day 4 to day 6. In queen bee larvae, these four miRNAs showed slightly different expression patterns, and the expression levels of miRNAs including ame-miR-12, ame-miR-263, and ame-miR-277 targeting gene JH acid methyltransferase increased from day 4 to day 5 yet decreased from day 5 to day 6; however, ame-miR-283 targeting JH-responsive metabolic enzymes of AMP-binding enzyme had an increased expression from day 4 to day 5 and then had similar expression levels or a slight increase in expression from day 5 to day 6. When comparing the expression of these miRNAs between worker and queen bee larvae, we can hypothesize that these JH-related miRNAs play critical roles in worker larvae at day 4 but seem to have greater influence on queen bee development at day 5.
4 Discussion
Several aspects including ecdysone, juvenile hormone (JH), TOR pathway, epigenetic modification, royalactin, and insulin have been reported to function in caste determination of worker and queen bee (Capella and Hartfelder 1998; Hartfelder et al. 2015; Kamakura 2011; Kucharski et al. 2008; Leimar et al. 2012; Patel et al. 2007; Pinto et al. 2002; Wheeler et al. 2006). We previously showed that miRNAs in jellies could influence the caste determination of worker and queen bee larvae (Guo et al. 2013). Here, we have profiled the small RNAs of fourth to sixth day worker and queen larvae using deep sequencing and qRT-PCR to explore the mechanism of caste determination and found that 53 known miRNAs were expressed in the early and critical caste developmental stages of bees, a number of which were shown higher in either queen or worker larval bees than the others, respectively. The qPCR results further demonstrated the dynamic expression patterns of those miRNAs along with caste development of queen and worker bees. Three JH-related/responsive genes were reported to play important roles in queen development and were upregulated in the queen bee (Barchuk et al. 2007; Capella and Hartfelder 1998; Hay et al. 1995; Li et al. 2013). Juvenile hormone acid methyltransferase (JHAMT) is an enzyme involved in the final steps of JH biosynthesis in bee, and its expression level was found to be highly correlated with the rates of JH biosynthesis (Li et al. 2013). When analyzed with qPCR from early larval stage to initial pupal stage (day 2, day 3, day 4, and day 5), our group has shown that JHAMT was more abundant in queens than that in workers in almost all the stages we analyzed, and it was most highly expressed in day 5 queens which was about 100 times higher than that in workers at the same stage (Li et al. 2013). By miRNA target analysis, we found that JHAMT could be regulated by miRNA ame-miR-263, ame-miR-263b, ame-miR-277, and ame-miR-33, and all these miRNAs were 1.3∼4.7 times more highly expressed in workers than that in queens by small RNA sequencing. The additional qPCR analysis for ame-miR-263 and ame-miR-277 in day 4, day 5, and day 6 further confirmed their overexpression in workers and also showed that both miRNAs were highest in day 4 workers, indicating that the highest expression of these miRNAs may to greatest extent repress the expression of JHAMT and subsequently limit the production of JH in day 4, and principally commit larvae bee to develop toward workers. Ame-miR-277 also targets gene LOC726040 (JH‐responsive metabolic enzymes of AMP‐binding enzyme, XM_001121814), which was reported significantly higher in queen larvae than that in workers and could be induced in worker larvae when treated with JH (Barchuk et al. 2007). Other miRNAs that could target LOC726040 include ame-miR-283 and ame-miR-31a, which were found to be higher in workers than ones in queens as well. Another JH-responsive gene is inhibitor of apoptosis 2 (IAP2, XM_396819) that could be targeted by ame-miR-12 and ame-miR-33 (Capella and Hartfelder 1998; Hay et al. 1995). This suggests that miRNAs and their targeted mRNAs co-regulate the caste differentiation of workers and queens.
In addition to the JH-related miRNAs, some other miRNAs including miRNA let-7, miR-100, miR-125, and miR-184 may play important functions in caste and development of bees as well. The loss of one or multiple of them in Drosophila induced the adult behavioral deficiencies in oviposition, fertility, and oogenesis (Iovino et al. 2009; Sokol et al. 2008). Since there are obverse discrepancies between workers and queens in adult behaviors such as flight, motility, oviposition, and fertility, we could hypothesize that miRNA let-7, miR-100, and miR-184 may play crucial roles in the caste switch in bees as JH-related miRNAs.
Our study has demonstrated the dynamic expression of miRNAs in the three key stages of bee caste differentiation and the distinct expression patterns of miRNAs between the two caste sisters; nevertheless, there are two limitations. First, the intervals of day 4, day 5, and day 6 are very crucial time points to investigate the miRNA variation in the differentiation of caste development; however, the additional sampling time points earlier from day 1 to day 3 and later after day 6 will be of worth to expression profile as well and compare with the three sampling time points we took. Second, no functional test of miRNAs we proposed to be relevant to the caste differentiation was performed in the current study. Therefore, the future coming expression profiling of wide stage span and functional study of miRNAs needs to be investigated.
5 Conclusions
We have systematically profiled the dynamic expression of small RNAs in worker and queen bee larvae by high-throughput sequencing and qPCR and have found a number of miRNAs that are differentially expressed between worker and queen larvae. Specific expression patterns and large discrepancies across small RNA profiles between worker and queen larvae indicate that small RNAs are potentially related to the differential development of worker and queen bee larvae. This study provides helpful information for exploring the caste switch between worker and queen bees and provides a solid foundation for investigating caste mechanisms in eusocial insects.
References
Audic, S., Claverie, J.-M. (1997) The significance of digital gene expression profiles. Genome Res 7(10), 986–995
Barchuk, A.R., Cristino, A.S., Kucharski, R., Costa, L.F., Simoes, Z.L., Maleszka, R. (2007) Molecular determinants of caste differentiation in the highly eusocial honeybee Apis mellifera. BMC Dev. Biol 7(70), 1–19
Bartel, D.P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2), 281–297
Betel, D., Wilson, M., Gabow, A., Marks, D.S., Sander, C. (2008) The microRNA.org resource: targets and expression. Nucleic Acids Res 36(Database issue), D149–153
Biryukova, I., Asmar, J., Abdesselem, H., Heitzler, P. (2009) Drosophila mir-9a regulates wing development via fine-tuning expression of the LIM only factor, dLMO. Dev Biol. 327(2), 487–496
Bomtorin, A.D., Mackert, A., Rosa, G.C., Moda, L.M., Martins, J.R., Bitondi, M.M., Hartfelder, K., Simoes, Z.L. (2014) Juvenile hormone biosynthesis gene expression in the corpora allata of honey bee (Apis mellifera L.) female castes. PLoS One 9(1), e86923
Brennecke, J., Hipfner, D.R., Stark, A., Russell, R.B., Cohen, S.M. (2003) bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113(1), 25–36
Cameron, R.C., Duncan, E.J., Dearden, P.K. (2013) Biased gene expression in early honeybee larval development. BMC Genomics 14(903), 12
Capella, I.C., Hartfelder, K. (1998) Juvenile hormone effect on DNA synthesis and apoptosis in caste-specific differentiation of the larval honey bee (Apis mellifera L.) ovary. J. Insect Physiol 44(5–6), 385–391
Chen, X., Yu, X., Cai, Y., Zheng, H., Yu, D., Liu, G., Zhou, Q., Hu, S., Hu, F. (2010) Next-generation small RNA sequencing for microRNAs profiling in the honey bee Apis mellifera. Insect Mol. Biol. 19(6), 799–805
Cheng, D., Meng, M., Peng, J., Qian, W., Kang, L., Xia, Q. (2014) Genome-wide comparison of genes involved in the biosynthesis, metabolism, and signaling of juvenile hormone between silkworm and other insects. Genet. Mol. Biol. 37(2), 444–459
Consortium, H.G.S. (2006) Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443(7114), 931–949
Dedej, S., Hartfelder, K., Aumeier, P., Rosenkranz, P., Engels, W. (1998) Caste determination is a sequential process: effect of larval age at grafting on ovariole number, hind leg size and cephalic volatiles in the honey bee (Apis mellifera carnica). J. Apic. Res. 37(3), 183–190
Escalier, D. (2008) Knockout mice in the service of reproduction. Gynecol. Obstet. Fertil. 36(12), 1224–1229
Evans, J.D., Wheeler, D.E. (1999) Differential gene expression between developing queens and workers in the honey bee, Apis mellifera. Proc. Natl. Acad. Sci. U S A 96(10), 5575–5580
Evans, J.D., Wheeler, D.E. (2001) Expression profiles during honeybee caste determination. Genome Biol. 2(1). RESEARCH0001
Farh, K.K., Grimson, A., Jan, C., Lewis, B.P., Johnston, W.K., Lim, L.P., Burge, C.B., Bartel, D.P. (2005) The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science 310(5755), 1817–1821
Greenberg, J.K., Xia, J., Zhou, X., Thatcher, S.R., Gu, X., et al. (2012) Behavioral plasticity in honey bees is associated with differences in brain microRNA transcriptome. Genes Brain Behav. 11(6), 660–670
Grun, D., Wang, Y.L., Langenberger, D., Gunsalus, K.C., Rajewsky, N. (2005) microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput. Biol. 1(1), e13
Guo, X., Su, S., Skogerboe, G., Dai, S., Li, W., et al. (2013) Recipe for a busy bee: microRNAs in Honey Bee caste determination. PLoS One 8(12), e81661
Hartfelder, K., Guidugli-Lazzarini, K.R., Cervoni, M.S., Santos, D.E., Humann, F.C. (2015) Old threads make new tapestry—Rewiring of signalling pathways underlies caste phenotypic plasticity in the honey bee Apis mellifera L. Adv. Insect Physiol. 48, 1–36
Hay, B.A., Wassarman, D.A., Rubin, G.M. (1995) Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell 83(7), 1253–1262
Hipfner, D.R., Weigmann, K., Cohen, S.M. (2002) The bantam gene regulates Drosophila growth. Genetics 161(4), 1527–1537
Hong, X., Luense, L.J., McGinnis, L.K., Nothnick, W.B., Christenson, L.K. (2008) Dicer1 is essential for female fertility and normal development of the female reproductive system. Endocrinology 149(12), 6207–6212
Iovino, N., Pane, A., Gaul, U. (2009) miR-184 has multiple roles in Drosophila female germline development. Dev. Cell 17(1), 123–133
Jiang, P., Wu, H., Wang, W., Ma, W., Sun, X., Lu, Z. (2007) MiPred: classification of real and pseudo microRNA precursors using random forest prediction model with combined features. Nucleic Acids Res. 35(suppl 2), W339–W344
Kamakura, M. (2011) Royalactin induces queen differentiation in honeybees. Nature 473(7348), 478–483
Kucharski, R., Maleszka, J., Foret, S., Maleszka, R. (2008) Nutritional control of reproductive status in honeybees via DNA methylation. Science 319(5871), 1827–1830
Kuhn, R.M., Karolchik, D., Zweig, A.S., Wang, T., Smith, K.E., et al. (2009) The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 37(Database issue), D755–761
Leimar, O., Hartfelder, K., Laubichler, M.D., Page Jr., R.E. (2012) Development and evolution of caste dimorphism in honeybees - a modeling approach. Ecol. Evol. 2(12), 3098–3109
Li, S.C., Tang, P., Lin, W.C. (2007) Intronic microRNA: discovery and biological implications. DNA Cell Biol. 26(4), 195–207
Li, R., Li, Y., Kristiansen, K., Wang, J. (2008) SOAP: short oligonucleotide alignment program. Bioinformatics 24(5), 713–714
Li, W., Huang, Z.Y., Liu, F., Li, Z., Yan, L., Zhang, S., Chen, S., Zhong, B., Su, S. (2013) Molecular cloning and characterization of juvenile hormone acid methyltransferase in the honey bee, Apis mellifera, and its differential expression during caste differentiation. PLoS One 8(7), e68544
Liu, F., Peng, W., Li, Z., Li, W., Li, L., et al. (2012) Next-generation small RNA sequencing for microRNAs profiling in Apis mellifera: comparison between nurses and foragers. Insect Mol. Biol. 21(3), 297–303
Luense, L.J., Carletti, M.Z., Christenson, L.K. (2009) Role of Dicer in female fertility. Trends Endocrinol. Metab. 20(6), 265–272
Moberg, K.H., Hariharan, I.K. (2003) Big things from a little RNA. Trends Cell Biol. 13(9), 455–457
Moritz, R.F., Lattorff, H.M., Neumann, P., Kraus, F.B., Radloff, S.E., Hepburn, H.R. (2005) Rare royal families in honeybees, Apis mellifera. Naturwissenschaften 92(10), 488–491
Page Jr., R.E., Peng, C.Y. (2001) Aging and development in social insects with emphasis on the honey bee, Apis mellifera L. Exp Gerontol. 36(4–6), 695–711
Patel, A., Fondrk, M.K., Kaftanoglu, O., Emore, C., Hunt, G., Frederick, K., Amdam, G.V. (2007) The making of a queen: TOR pathway is a key player in diphenic caste development. PLoS One 2(6), e509
Pinto, L.Z., Hartfelder, K., Bitondi, M.M.G., Simoes, Z.L.P. (2002) Ecdysteroid titers in pupae of highly social bees relate to distinct modes of caste development. J. Insect Physiol. 48(8), 783–790
Rembold, H., Czoppelt, C., Rao, P.J. (1974) Effect of juvenile hormone treatment on caste differentiation in the honeybee, Apis mellifera. J. Insect Physiol. 20(7), 1193–1202
Rodriguez, A., Griffiths-Jones, S., Ashurst, J.L., Bradley, A. (2004) Identification of mammalian microRNA host genes and transcription units. Genome Res. 14(10A), 1902–1910
S. C., S. S., X. L. (2002) An introduction to high-yielding royal jelly production methods in China. Bee World 83,69-77
Shi, Y.Y., Wu, X.B., Huang, Z.Y., Wang, Z.L., Yan, W.Y., Zeng, Z.J. (2012) Epigenetic modification of gene expression in honey bees by heterospecific gland secretions. PLoS One 7(8), e43727
Shi, Y., Zheng, H., Pan, Q., Wang, Z., Zeng, Z. (2015) Differentially expressed microRNAs between queen and worker larvae of the honey bee (Apis mellifera ). Apidologie 46(1), 35–45
Shuel, R.W., Dixon, S.E. (1960) The early establishment of dimorphism in the female honeybee, Apis mellifera L. Insectes Soc. 7(3), 265–282
Sokol, N.S., Xu, P., Jan, Y.-N., Ambros, V. (2008) Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes & Devt 22(12), 1591–1596
Wang, X., Zhang, J., Li, F., Gu, J., He, T., Zhang, X., Li, Y. (2005) MicroRNA identification based on sequence and structure alignment. Bioinformatics 21(18), 3610–3614
Weaver, D., Anzola, J., Evans, J., Reid, J., Reese, J., et al. (2007) Computational and transcriptional evidence for microRNAs in the honey bee genome. Genome Biol. 8(6), R97
Wheeler, D.E., Buck, N., Evans, J.D. (2006) Expression of insulin pathway genes during the period of caste determination in the honey bee, Apis mellifera. Insect Mol. Biol. 15(5), 597–602
Yang, Y., Xu, S., Xia, L., Wang, J., Wen, S., Jin, P., Chen, D. (2009) The bantam microRNA is associated with drosophila fragile X mental retardation protein and regulates the fate of germline stem cells. PLoS Genet. 5(4), e1000444
Ying, S.Y., Lin, S.L. (2006) Current perspectives in intronic micro RNAs (miRNAs). J Biomed. Sci. 13(1), 5–15
Zondag, L., Dearden, K.P., Wilson, J.M. (2012) Deep sequencing and expression of microRNAs from early honeybee (Apis mellifera) embryos reveals a role in regulating early embryonic patterning. BMC Evol. Biol. 12(21), 1–13
Acknowledgments
The deep sequencing was carried out at the Beijing Genome Institute (http://www.genomics.cn/index.php). We thank Shuanjin Dai, Yunjuan Hu, and Qin Chen for their help in research. We thank Drs. Aung Si and Erna Forgo, who read and improved the manuscript.
Author’s contributions
RC, SZ, and SC conceived and designed the study. XG, SS, and SG participated in the study design and drafted the manuscript. XG and SS performed the experiment and analysis. WL and ZL participated in the data analysis. RC, SZ, SC, and SS contributed reagents/materials/analysis tool. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Manuscript editor: Klaus Hartfelder
Expression différentielle des micro-ARN en lien avec la différenciation des castes chez l'abeille Apis mellifera
Apidae / petits ARN / développement larvaire / détermination des castes
Differentielle Expression von miRNAs mit Bezug zur Kastendifferenzierung in der Honigbiene, Apis mellifera
Honigbiene / kurze RNAs / Larvenentwicklung / Kastenentwicklung
Xiangqian Guo and Songkun Su contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
The qPCR primers used in this study. (PDF 82 kb)
ESM 2
Expression levels of miRNAs corresponding to known miRNAs. (PDF 70 kb)
ESM 3
List and annotation of the novel bee miRNAs. (XLS 344 kb)
ESM 4
Pairs of novel miRNAs and their miRNAs. (XLS 50 kb)
ESM 5
Summary of novel miRNAs and their miRNAs. (PDF 48 kb)
ESM 6
The three JH genes and miRNAs targeting them. (PDF 50 kb)
ESM 7
qPCR results of known miRNAs. (PDF 501 kb)
ESM 8
qPCR results of novel miRNAs. (PDF 690 kb)
ESM 9
qPCR results of JH relative miRNAs. (PDF 401 kb)
Rights and permissions
About this article
Cite this article
Guo, X., Su, S., Geir, S. et al. Differential expression of miRNAs related to caste differentiation in the honey bee, Apis mellifera . Apidologie 47, 495–508 (2016). https://doi.org/10.1007/s13592-015-0389-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13592-015-0389-3