Abstract
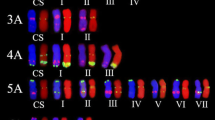
Triticum boeoticum (2n = 2x = 14, AbAb) contains beneficial traits for common wheat improvement. The discrimination of Ab-genome chromosomes from A-genome chromosomes is an important step in gene transfer from T. boeoticum to common wheat. In this study, fluorescence in situ hybridization (FISH) analysis using nine oligonucleotide probes revealed high divergence between chromosomes of the common wheat germplasm Crocus and T. boeoticum accession G52. The combination of Oligo-pTa535-HM and Oligo-pSc119.2-HM can differentiate Ab and A chromosomes within homologous groups 2, 4, 5, and 6; chromosomes 2Ab and 6Ab can be identified by using (ACT)7, (CTT)7, and (GAA)7. The probes Oligo-pTa713 and (ACT)7 can be utilized for the identification of chromosomes 1Ab and 3Ab, respectively. Probes (CAG)7 and (CAC)7 can be applied in the identification of 7Ab. Moreover, probe combinations consisting of Oligo-pTa535-HM and (AAC)7 with (ACT)7 or (CTT)7 and of Oligo-pTa535-HM and Oligo-pTa713 with (CAC)7 or (CTT)7 will help discriminate the Ab-genome chromosomes of T. boeoticum. These probes are being used as potential markers to select common wheat Crocus-T. boeoticum G52 alien chromosome lines. Moreover, FISH patterns are highly divergent between Ab- and A-genome chromosomes, indicating that obvious chromosome structural variations arose during wheat evolution.




Similar content being viewed by others
References
Adonina IG, Goncharov NP, Badaeva ED, Sergeeva EM, Petrash NV, Salina EA (2015) (GAA)n microsatellite as an indicator of the a genome reorganization during wheat evolution and domestication. Comp Cytogenet 9:533–547
Anker CC, Niks RE (2001) Prehaustorial resistance to wheat leaf rust in Triticum monococcum (s.s.). Euphytica 117:209–215
Badaeva ED, Amosova AV, Goncharov NP, Macas J, Ruban AS, Grechishnikova IV, Zoshchuk SA, Houben A (2015) A set of cytogenetic markers allows the precise identification of all A-genome chromosomes in diploid and polyploid wheat. Cytogenet Genome Res 146:71–79
Badaeva ED, Ruban AS, Zoshchuk SA, Surzhikov SA, Knüpffer H, Kilian B (2016) Molecular cytogenetic characterization of Triticum timopheevii chromosomes provides new insight on genome evolution of T. zhukovskyi. Plant Syst Evol 302:943–956
Beliveau BJ, Joyce EF, Apostolopoulos N, Yilmaz F, Fonseka CY, McCole RB, Changa YM, Lia JB, Senaratnea TN, Williamsa BR, Rouillard J, Wu C (2012) Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc Natl Acad Sci U S A 109:21301–21306
Carmona FE, Bustos AD, Jouve N, Cuadrado A (2013) Cytogenetic diversity of SSR motifs within and between Hordeum species carrying the H genome: H. vulgare L. and H. bulbosum L. Theor Appl Genet 126:949–961
Castagna R, Borghi B, Huen M, Salamini F (1995) Integrated approach to einkorn wheat breeding. In Hulled wheats. Promoting the conservation and use of underutilized and neglected crops 4. In: Proceedings Wrst Intl. workshop on Hulled wheat, Castelvecchio Pascoli, Tuscany, Italy. IPGRI, Rome, Italy, pp 21–22
Contento A, Hslop-Harrison JS, Schwarzacher T (2005) Diversity of a major repetitive DNA sequence in diploid and polyploid Triticeae. Cytogenet Genome Res 109:34–42
Cuadrado A, Jouve N (2002) Evolutionary trends of different repetitive DNA sequences during speciation in the genus Secale. J Hered 93:339–345
Cuadrado A, Schwarzacher T (1998) The chromosomal organization of simple sequence repeats in wheat and rye genomes. Chromosoma 107:587–594
Cuadrado A, Cardoso M, Jouve N (2008) Physical organisation of simple sequence repeats (SSRs) in Triticeae: structural, functional and evolutionary implications. Cytogenet Genome Res 120:210–219
Danilova TV, Friebe B, Gill BS (2012) Single-copy gene fluorescence in situ hybridization and genome analysis: Acc-2 loci mark evolutionary chromosomal rearrangements in wheat. Chromosoma 121:597–611
Du P, Zhuang LF, Wang YZ, Yuan L, Wang Q, Wang DR, Dawadondup, Tan LJ, Shen J, Xu HB, Zhao H, Chu CG, Qi ZJ (2017) Development of oligonucleotides and multiplex probes for quick and accurate identification of wheat and Thinopyrum bessarabicum chromosomes. Genome 60:93–103
Dvorák J, McGuire PE, Cassidy B (1988) Apparent sources of the a genome of wheat inferred from polymorphism in abundance and restriction fragment length of repeated nucleotide sequences. Genome 30:680–689
Flavell RB (1986) Repetitive DNA and chromosome evolution in plants. Philos Trans R Soc Lond Ser B Biol Sci 312:227–242
Flavell RB, Bennett MD, Smith JB, Smith DB (1974) Genome size and the proportion of repeated nucleotide sequence DNA in plants. Biochem Genet 12:257–269
Fox SE, Geniza M, Hanumappa M, Naithani S, Sullivan C, Preece J, Tiwari VK, Elser J, Leonard JM, Sage A, Gresham C, Kerhornou A, Bolser D, McCarthy F, Kersey P, Lazo GR, Jaiswal P (2014) De novo transcriptome assembly and analyses of gene expression during photomorphogenesis in diploid wheat Triticum monococcum. PLoS One 9:e96855
Fu SL, Chen L, Wang YY, Li M, Yang ZJ, Qiu L, Yan BJ, Ren ZL, Tang ZX (2015) Oligonucleotide probes for ND-FISH analysis to identify rye and wheat chromosomes. Sci Rep 5:10552
Grewal S, Hubbart-Edwards S, Yang C, Scholefield D, Ashling S, Burridge A, Wilkinson PA, King IP, King J (2018) Detection of T. urartu introgressions in wheat and development of a panel of interspecific introgression lines. Front. Plant Sci 9:1565
Han YH, Zhang T, Thammapichai P, Weng YQ, Jiang JM (2015) Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics 200:771–779
Hao M, Luo JT, Zhang LQ, Yuan ZW, Yang YW, Wu M, Chen WJ, Zheng YL, Zhang HG, Liu DC (2013) Production of hexaploid triticale by a synthetic hexaploid wheat-rye hybrid method. Euphytica 193:347–357
Harjit Singh, Grewal TS, Dhaliwal HS, Pannu PPS, Bagga PPS (1998) Sources of leaf rust and stripe rust resistance in wild relatives of wheat. Crop Improv 25:26–33
He Q, Cai Z, Hu T, Liu H, Bao C, Mao W, Jin W (2015) Repetitive sequence analysis and karyotyping reveals centromere-associated DNA sequences in radish (Raphanus sativus L.). BMC Plant Biol 15:105
Heslop-Harrison JS, Schmidt T (1998) Genomes, genes and junk: the large-scale organization of plant genomes. Trends Plant Sci 3:195–199
Heun M, Schäfer-Pregl R, Klawan D, Castagna R, Accerbi M, Borghi B, Salamini F (1997) Site of einkorn wheat domestication identified by DNA fingerprinting. Science 278:1312–1314
Hovhannisyan NA, Dulloo MH, Yesayan AH, Knüpffer H, Amri A (2011) Tracking of powdery mildew and leaf rust resistance genes in Triticum boeoticum and T. urartu, wild wheat relatives of common wheat. Czech J Genet Plant Breed 47:45–57
Johnson BL, Dahliwal HS (1976) Reproductive isolation of Triticum boeoticum and Triticum urartu and the origin of the tetraploid wheats. Am J Bot 63:1088–1094
Komuro S, Endo R, Shikata K, Kato A (2013) Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome 56:131–137
Kubaláková M, Kovářová P, Suchánková P, Číhalíková J, Bartoš J, Lucretti S, Watanabe N, Kianian SF, Doležel J (2005) Chromosome sorting in tetraploid wheat and its potential for genome analysis. Genetics 170:823–829
Lebedeva TV, Peusha HO (2006) Genetic control of the wheat Triticum monococcum L. resistance to powdery mildew. Genetika 42:71–77
Li GR, Gao D, Zhang HG, Li JB, Wang HJ, La SX, Ma JW, Yang ZJ (2016) Molecular cytogenetic characterization of Dasypyrum breviaristatum chromosomes in wheat background revealing the genomic divergence between Dasypyrum species. Mol Cytogenet 9:6
Li HY, Liu XJ, Zhang MH, Feng Z, Liu DC, Ayliffe M, Hao M, Ning SZ, Yuan ZW, Yan ZH, Chen XJ, Zhang LQ (2018) Development and identification of new synthetic T. turgidum–T. monococcum amphiploids. Plant Genet Resour C 16(6):555–563
Ling HQ, Zhao S, Liu D, Wang J, Sun H, Zhang C, Fan H, Li D, Dong L, Tao Y, Gao C, Wu H, Lii Y, Cui Y, Guo X, Zheng S, Wang B, Yu K, Liang Q, Yang W, Lou X, Chen J, Feng M, Jian J, Zhang X, Luo G, Jiang Y, Liu J, Wang Z, Sha Y, Zhang B, Wu H, Tang D, Shen Q, Xue P, Zou S, Wang X, Liu X, Wang F, Yang Y, Zn X, Dong Z, Zhang K, Zhang X, Luo MC, Dvorak J, Tong Y, Wang J, Yang H, Li Z, Wang D, Zhang A, Wang J (2013) Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 496:87–90
Ling HQ, Ma B, Shi X, Liu H, Dong L, Sun H, Cao Y, Gao Q, Zheng S, Li Y, Yu Y, Du H, Qi M, Li Y, Lu H, Yu H, Cui Y, Wang N, Chen C, Wu H, Zhao Y, Zhang J, Li Y, Zhou W, Zhang B, Hu W, van Eijk MJT, Tang J, Witsenboer HMA, Zhao S, Li Z, Zhang A, Wang D, Liang C (2018) Genome sequence of the progenitor of wheat a subgenome Triticum urartu. Nature 557:424–428
Liu XJ, Zhang MH, Liu X, Li HY, Hao M, Ning SZ, Yuan ZW, Liu DC, Wu BH, Chen XJ, Chen WJ, Zhang LQ (2018) Molecular cytogenetic identification of newly synthetic Triticum kiharae with high resistance to stripe rust. Genet Resour Crop Evol 65(6):1725–1732
McIntyre CL, Pereira S, Moran LB, Appels R (1990) New Secale cereale (rye) DNA derivatives for the detection of rye chromosome segments in wheat. Genome 33:635–640
Megyeri M, Farkas A, Varga M, Kovács G, Molnár-Láng M, Molnár I (2012) Karyotypic analysis of Triticum monococcum using standard repetitive DNA probes and simple sequence repeats. Acta Agron Hung 60:87–95
Megyeri M, Mikó P, Farkas A, Molnár-Láng M, Molnár I (2017) Cytomolecular discrimination of the am chromosomes of Triticum monococcum and the A chromosomes of Triticum aestivum using microsatellite DNA repeats. J Appl Genet 58:67–70
Mehrotra S, Goyal V (2014) Repetitive sequences in plant nuclear DNA: types, distribution, evolution and function. Genom Proteom Bioinf 12:164–171
Michikawa A, Yoshida K, Okada M, Sato K, Takumi S (2019) Genome-wide polymorphisms from RNA sequencing assembly of leaf transcripts facilitate phylogenetic analysis and molecular marker development in wild einkorn wheat. Mol Gen Genomics 294:1327–1341
Mikó P, Megyeri M, Farkas A, Molnár I, Molnár-Láng M (2015) Molecular cytogenetic identification and phenotypic description of a new synthetic amphiploid, Triticum timococcum (AtAtGGAmAm). Genet Res Crop Evol 62:55–66
Mirzaghaderi G, Houben A, Badaeva E (2014) Molecular-cytogenetic analysis of Aegilops triuncialis and identification of its chromosomes in the background of wheat. Mol Cytogenet 7:91
Molnár I, Linc G, Dulai S, Nagy ED, Molnár-Láng M (2007) Ability of chromosome 4H to compensate for 4D in response to drought stress in a newly developed and identified wheat-barley 4H(4D) disomic substitution line. Plant Breed 126:369–374
Pedersen C, Langridge P (1997) Identification of the entire chromosome complement of bread wheat by two-colour FISH. Genome 40:589–593
Pedersen C, Rasmussen SK, Linde-Laursen I (1996) Genome and chromosome identification in cultivated barley and related species of the Triticeae (Poaceae) by in situ hybridization with the GAA-satellite sequence. Genome 39:93–104
Rogers WJ, Miller TE, Payne PI, Seekings JA, Sayers EJ, Holt LM, Law CN (1997) Introduction to bread wheat (Triticum aestivum L.) and assessment for bread-making quality of alleles from T. boeoticum Boiss. Ssp. thaoudar at Glu-A1 encoding two high-molecular-weight subunits of glutenin. Euphytica 93:19–29
Schneider A, Linc G, Molnár-Láng M (2003) Fluorescence in situ hybridization polymorphism using two repetitive DNA clones in different cultivars of wheat. Plant Breed 122:396–400
Sepsi A, Molnár I, Szalay D, Molnár-Láng M (2008) Characterization of a leaf rust resistant wheat–Thinopyrum ponticum partial amphiploid BE-1 using sequential multicolor GISH and FISH. Theor Appl Genet 116:825–834
Takumi S, Nasuda S, Liu YG, Tsunewaki K (1993) Wheat phylogeny determined by RFLP analysis of nuclear DNA. 1. Einkorn wheat. Jpn J Genet 68:73–79
Tang ZX, Yang ZJ, Fu SL (2014) Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J Appl Genet 55:313–318
Tang SY, Qiu L, Xiao ZQ, Fu SL, Tang ZX (2016) New oligonucleotide probes for ND-FISH analysis to identify barley chromosomes and to investigate polymorphisms of wheat chromosomes. Genes 7:118
Tautz D (1989) Hypervariability of simple sequences as a general source for polymorphic markers. Nucleic Acids Res 17:6463–6471
Vasu K, Harjit-Singh, Chhuneja P, Singh S, Dhaliwal HS (2000) Molecular tagging of Karnal bunt resistance genes of Triticum monococcum transferred to Triticum aestivum L. Crop Improv 27:33–42
Xiao ZQ, Tang SY, Qiu L, Tang ZX, Fu SL (2017) Oligonucleotides and ND-FISH displaying different arrangements of tandem repeats and identification of Dasypyrum villosum chromosomes in wheat backgrounds. Molecules 22:973
Zhang RQ, Zhang MY, Wang XE, Chen PD (2014) Introduction of chromosome segment carrying the seed storage protein genes from chromosome 1V of Dasypyrum villosum showed positive effect on bread-making quality of common wheat. Theor Appl Genet 127:523–533
Zhao LB, Ning SZ, Yu JJ, Hao M, Zhang LQ, Yuan ZW, Zheng YL, Liu DC (2016) Cytological identification of an Aegilops variabilis chromosome carrying stripe rust resistance in wheat. Breed Sci 66:522–529
Zhao LB, Ning SZ, Yi YJ, Zhang LQ, Yuan ZW, Wang JR, Zheng YL, Hao M, Liu DC (2018) Fluorescence in situ hybridization karyotyping reveals the presence of two distinct genomes in the taxon Aegilops tauschii. BMC Genomics 19:3
Funding
This research was financially supported by the National Natural Science Foundation of China (31671682, 31671689).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by: Izabela Pawłowicz
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Fig. S1
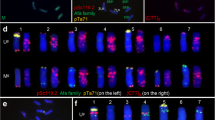
Photomicrographs showing the distribution of several probes on metaphase chromosomes of the T. boeoticum accession G52. a Oligo-pTa535-HM (green), b (CAG)7 (green), c (GAA)7 (green), d (AAC)7 (red), e (CTT)7 (red), f Oligo-pTa713 (red), g (CAC)7 (green), h (ACT)7 (red). (PNG 2118 kb)
Fig. S2
Photomicrographs showing the distribution of several probes on metaphase chromosomes of the common wheat germplasm Crocus. a Oligo-pTa535-HM (green), Oligo-pSc119.2-HM (red), b (CAG)7 (green), c (GAA)7 (green), d (AAC)7 (red), e (CTT)7 (red), f (ACT)7 (red), g Oligo-pTa713 (red), h (CAC)7 (green). (PNG 2468 kb)
Rights and permissions
About this article
Cite this article
Feng, Z., Zhang, M., Liu, X. et al. FISH karyotype comparison between Ab- and A-genome chromosomes using oligonucleotide probes. J Appl Genetics 61, 313–322 (2020). https://doi.org/10.1007/s13353-020-00555-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-020-00555-7