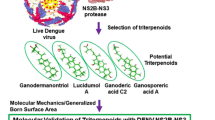
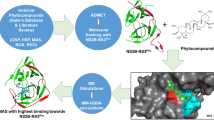
Abstract
Dengue virus (DENV) has emerged as an increasing fitness problem in the world for which no specialized drug is available. The non-structural protein NS3 protease of DENV has already been recognized as a potential therapeutic target for the discovery and development of novel antiviral agents against DENV infections. In this study, we employed the virtual screening technique to explore the potent inhibitors of DENV NS2B/NS3pro from Traditional Chinese Medicine (TCM) database. Total 200 inhibitors from TCM against DENV NS3pro were screened and only five TCM compounds like eriodictyol 7-O-glucuronide, luteolin 8-C-beta-glucopyranoside, (-)-epicatechin-3-O-gallate, 6-O-trans-p-coumaroylgeniposide and luteolin-7-O-glucoside were selected for further analysis which showed binding energies, −7.000, −7.380, −7.380, −7.440 and −7.440 kcal/mol, respectively. The findings of this study suggest that these five TCM compounds can be considered as potent inhibitors for DENV NS2B/NS3pro for the development of anti-dengue drugs.

Similar content being viewed by others
References
Anand P, Nagarajan D, Mukherjee S, Chandra N. PLIC: protein–ligand interaction clusters. Database, 2014, bau029.
Anciotti RS, Gubler DJ, Trent DW. Molecular evolution and phylogeny of dengue-4 viruses. J Gen Virol. 1997;78:2279–86.
Anderson CR, Downs WG, Hill AE. Isolation of dengue virus from a human being in Trinidad. Science. 1956;124:224–5.
Arias CF, Preugschat F, Strauss JH. Dengue 2 virus NS2B and NS3 form a stable complex that can cleave NS3 within the helicase domain. Virology. 1993;193:888–99.
Berman HM, Rose PW, Dutta S, Zardecki C, Prlić A. The Protein Data Bank: overview and tools for drug discovery. In: Multifaceted roles of crystallography in modern drug discovery. 2015. pp. 93–106.
Chambers TJ, Nestorowicz A, Amberg SM, Rice CM. Mutagenesis of the yellow fever virus NS2B protein: effects on proteolytic processing, NS2B-NS3 complex formation, and viral replication. J Virol. 1993;67:6797–807.
Chen CYC. TCM Database@ Taiwan: the world’s largest traditional Chinese medicine database for drug screening. PLoS One. 2011;6:e15939.
de Almeida H, Bastos IM, Ribeiro BM, Maigret B, Santana JM. New binding site conformations of the dengue virus NS2B-NS3 protease accessed by molecular dynamics simulation. PLoS One. 2013;8:e72402.
DeLano WL. Pymol: an open-source molecular graphics tool. CCP4 Newslett Protein Crystallogr. 2002;40:82–92.
Dwivedi VD, Tripathi IP, Mishra SK. In silico evaluation of inhibitory potential of triterpenoids from Azadirachta indica against therapeutic target of dengue virus, NS2B-NS3 protease. J Vector Borne Dis. 2016;53:156–161.
Falgout B, Pethel M, Zhang YM, Lai CJ. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus 651 nonstructural proteins. J Virol. 1991;65:2467–75.
Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11:480–96.
Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov. 2004;3:935–49.
Labbé CM, Rey J, Lagorce D, Vavruša M, Becot, J, et al. MTiOpenScreen: a web server for structure-based virtual screening. Nucleic Acids Res 2015; gkv306.
Laskowski RA, Swindells MB. LigPlot + : multiple ligand–protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51:2778–86.
Lee YK, Tan SK, Wahab HA, Rohana Y. Nonsubstrate based inhibitors of dengue virus serine protease: a molecular docking approach to study binding interactions between protease and inhibitors. Asia Pac J Mol Biol Biotechnol. 2007;15:53–9.
Lescar J, Luo D, Xu T, Sampath A, Lim SP, Canard B, Vasudevan SG. Towards the design of antiviral inhibitors against flaviviruses: the case for the multifunctional NS2B-NS3 protein from Dengue virus as a target. Antivir Res. 2008;80:94–101.
Luo D, Vasudevan S, Lescar J. The flavivirus NS2B–NS3 protease–helicase as a target for antiviral drug. Development. 2015;6:7.
Luo D, Xu T, Hunke C, Grüber G, Vasudevan SG, Lescar J. Crystal structure of the NS3 protease-helicase from dengue virus. J Virol. 2008;82:173–83.
Matusan AE, Pryor MJ, Davidson AD, Wright PJ. Mutagenesis of the dengue virus type 2 NS3 protein within and outside helicase motifs: effects on enzyme activity and virus replication. J Virol. 2001;75(20):9633–43.
Mustafa MS, Rasotgi V, Jain S, Gupta V. Discovery of fifth serotype of dengue virus (DENV-5): a new public health dilemma in dengue control. Med J Armed Forces India. 2015;71:67–70.
Natarajan S. NS2B-NS3 protease from flavivirus as a target for designing antiviral inhibitors against dengue virus. Genet Mol Biol. 2010;33:214–9.
Nitsche C, Behnam MM, Steuer C, Klein CD. Retro peptide-hybrids as selective inhibitors of the Dengue virus NS2B- NS2B-NS3 protease. Antivir Res. 2012;94:72–9.
Normile D. Surprising new dengue virus throws a spanner in disease control efforts. Science. 2013;342(6157):415.
Othman R, Kiat TS, Khalid N, Yusof R, Irene NE, Newhouse JS, et al. Docking of noncompetitive inhibitors into dengue virus type 2 protease: understanding the interactions with allosteric binding sites. J Chem Inf Model. 2008;48:1582–91.
Patil R, Das S, Stanley A, Yadav L, Sudhakar A, Varma AK. Optimized hydrophobic interactions and hydrogen bonding at the target-ligand interface leads the pathways of drug-designing. PLoS One. 2010;5:e12029.
Ranjit S, Kissoon N. Dengue hemorrhagic fever and shock syndromes*. Pediatr Crit Care Med. 2011;12:90–100.
Rigau-Pérez JG, Clark GG, Gubler DJ, Reiter P, Sanders EJ, Vorndam AV. Dengue and dengue haemorrhagic fever. Lancet. 1998;352(9132):971–7.
Rodenhuis-Zybert IA, Wilschut J, Smit JM. Dengue virus life cycle: viral and host factors modulating infectivity. Cell Mol Life Sci. 2010;67(16):2773–86.
Rose PW, Prlić A, Bi C, Bluhm WF, Christie CH, Dutta S, et al. The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nucleic Acids Res. 2015;43(D1):345–56.
Russell PK, Buescher EL, McCown JM, Ordoñez J. Recovery of dengue viruses from patients during epidemics in Puerto Rico and East Pakistan. Am J Trop Med Hyg. 1966;15:573–9.
Sabin AB, Schlesinger RW. Production of immunity to dengue with virus modified by propagation in mice. Science. 1945;101(2634):640–2.
Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M. PLIP: fully automated protein–ligand interaction profiler. Nucleic Acids Res 2015; gkv315.
Sarkar JK, Chatterjee SN, Chakravarty SK. Haemorrhagic fever in calcutta: some epidemiological observations. Indian J Med Res. 1964;52:651–9.
Simmons CP, Farrar JJ, Van VCN, Wills B. Dengue. N Engl J Med. 2012;366(15):1423–32.
Tiew KC, Dou D, Teramoto T, Lai H, Alliston KR, et al. Inhibition of dengue virus and west nile virus proteases by click chemistry-derived benz [d]isothiazol-3(2H)-one derivatives. Bioorg Med Chem. 2012;20:1213–21.
Tomlinson SM, Watowich SJ. Use of parallel validation high-throughput screens to reduce false positives and identify novel dengue NS2B-NS2B-NS3 protease inhibitors. Antivir Res. 2012;93:245–52.
Tsai TY, Chang KW, Chen CYC. IScreen: world’s first cloud-computing web server for virtual screening and de novo drug design based on TCM database@ Taiwan. J Comput Aided Mol Des. 2011;25:525–31.
Valle RP, Falgout B. Mutagenesis of the NS3 protease of dengue virus type 2. J Virol. 1998;72:624–32.
Velmurugan D, Mythily U, Rao K. Design and docking studies of peptide inhibitors as potential antiviral drugs for dengue virus ns2b/ns3 protease. Protein Pept Lett. 2014;21:815–27.
Wallace AC, Laskowski RA. Thornton JM LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–34.
Zhu Q, Bang TH, Ohnuki K, Sawai T, Sawai K, Shimizu K. Inhibition of neuraminidase by Ganoderma triterpenoids and implications for neuraminidase inhibitor design. Sci Rep 2015;5.
Acknowledgments
Authors acknowledge the support provided by Department of Biotechnology, D.D.U. Gorakhpur University, Gorakhpur.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dwivedi, V.D., Tripathi, I.P., Bharadwaj, S. et al. Identification of new potent inhibitors of dengue virus NS3 protease from traditional Chinese medicine database. VirusDis. 27, 220–225 (2016). https://doi.org/10.1007/s13337-016-0328-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13337-016-0328-6