Abstract
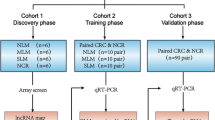
Recurrence, invasion, and metastasis are the major reasons of the low 5-year survival of hepatocellular carcinoma. However, the mechanisms of recurrence, invasion, and metastasis are still poll understood. Long noncoding RNAs (LncRNAs, >200 nt) have been demonstrated to play important roles in both tumor suppressive and oncogenic signaling pathways. Here, we employed the LncRNAs microarray technology to study the LncRNAs expression profiles at genome-wide in hepatocellular carcinoma (HCC) tissue samples with early recurrence (less than 1 year, with invasion and metastasis out of liver) and late recurrence (longer than 2 years, without invasion and metastasis out of liver), which had different recurrent/metastatic potentials, by using normal liver tissue as control to screen the dysregulated LncRNAs which are potentially involved in the recurrence, invasion, and metastasis process of HCC. Overall, 1170 LncRNAs were identified to differentially expressed between the early and late recurrence samples. These differentially expressed LncRNAs were further characterized by integrating examination of genomic context, co-expression network analysis, and gene ontology (GO) enrichment of their associated protein-coding genes. Furthermore, 15 LncRNAs selected randomly from top 50 differentially expressed LncRNAs were validated by quantitative PCR (qPCR) in cell lines MHCC97H and MHCC97L, which have exactly the same genetic background but with different invasion potentials. Meanwhile, the prognostic potential of three verified LncRNAs at cell line level was further validated in 59 HCC samples. Therefore, our results demonstrated that the aberrant expression of LncRNAs might be responsible for the HCC invasion and metastasis and provide fundamental information for further study the LncRNAs involved molecular mechanisms of the invasion and metastasis of HCC.








Similar content being viewed by others
References
Jemal A et al. Global cancer statistics. CA: Cancer J Clin. 2011;2:69–90.
Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA: Cancer J Clin. 2012;1:10–29.
Allemani C et al. Global surveillance of cancer survival 1995–2009: analysis of individual data for 25 676 887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet. 2014. doi:10.1016/S0140-6736(14)62038-9.
Rahbari NN, Mehrabi A, Mollberg NM, Müller SA, Koch M, Büchler MW, et al. Hepatocellular carcinoma: current management and perspectives for the future. Ann Surg. 2011;3:453–69.
Ng KM, Yan T, Black D, Chu FC, Morris DL. Prognostic determinants for survival after resection/ablation of a large hepatocellular carcinoma. HPB (Oxford). 2009;4:311–20.
Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;6:904–14.
Huang J-l, Zheng L, Hu Y-W. Characteristics of long noncoding RNA and its relation to hepatocellular carcinoma. Carcinogenesis. 2014;3:507–14.
Ernst C, Morton CC. Identification and function of long non-coding RNA. Front Cell Neurosci. 2013. doi:10.3389/fncel.2013.00168.
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;4:629–41.
Quagliata L et al. LncRNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2013;3:911–23.
Panzitt K et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;1:330–42.
Wang J et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;16:5366–83.
Yang F et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;5:1679–89.
Yuan J-H et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;5:666–81.
Wang F, Yuan J-H, Wang S-B, Yang F, Yuan S-X, Ye C, et al. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology. 2014;4:1278–90.
Yu W et al. Tumor suppressor long non-coding RNA, MT1DP is negatively regulated by YAP and Runx2 to inhibit FoxA1 in liver cancer cells. Cell Signal. 2014;12:2961–8.
Ørom UA et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;1:46–58.
Barabási A-L, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;2:101–13.
Dinger ME et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;9:1433–45.
Mercer TR et al. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010. doi:10.1186/1471-2202-11-14.
Carninci P et al. The transcriptional landscape of the mammalian genome. Science. 2005;5740:1559–63.
Wang X et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;7200:126–30.
Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;9:637–43.
Louro R et al. Conserved tissue expression signatures of intronic noncoding RNAs transcribed from human and mouse loci. Genomics. 2008;1:18–25.
Louro R, Smirnova AS, Verjovski-Almeida S. Long intronic noncoding RNA transcription: expression noise or expression choice? Genomics. 2009;4:291–8.
Trinklein ND et al. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;1:62–6.
Morris KV et al. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;11:e1000258.
Liu J et al. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell Online. 2012;11:4333–45.
Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;6:703–19.
Prensner JR et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;8:742–9.
Gupta RA et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;7291:1071–6.
Gutschner T, Hämmerle M, Diederichs S. MALAT1—a paradigm for long noncoding RNA function in cancer. J Mol Med. 2013;7:791–801.
Braconi C et al. Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma. Proc Natl Acad Sci. 2011;2:786–91.
Huang JF et al. Hepatitis B virus X protein (HBx)-related long noncoding RNA (LncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology. 2013;5:1882–92.
Yang F et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;6:1083–96.
Tsang WP, Kwok T. Riboregulator H19 induction of MDR1-associated drug resistance in human hepatocellular carcinoma cells. Oncogene. 2007;33:4877–81.
Zhu J et al. The long noncoding RNA expression profile of hepatocellular carcinoma identified by microarray analysis. PLoS One. 2014;9(7):e101707.
Kutter C et al. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet. 2012;7:e1002841.
Derrien T et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;9:1775–89.
Liu L et al. Maelstrom promotes hepatocellular carcinoma metastasis by inducing epithelial-mesenchymal transition by way of Akt/GSK-3β/Snail signaling. Hepatology. 2013;2:531–43.
Jiang D-K et al. Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat Genet. 2012;1:72–5.
Jiang J-H et al. An X-chromosomal association study identifies a susceptibility locus at Xq22. 1 for hepatitis B virus-related hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2013;6:586–95.
Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;3:155–9.
Sulzmaier FJ, Ramos JW. RSK isoforms in cancer cell invasion and metastasis. Cancer Res. 2013;20:6099–105.
Acknowledgments
This work is supported by the Key Clinical Specialty Discipline Construction Program of Fujian, P. R.C.; the Key Project of National Science and Technology of China (Grant no. 2012ZX10002010-001-006 and Grant no. 2012ZX10002016-013), the National Natural Science Foundation of China (Grant no. 31201008), the Key Project of Fujian Province (Grant no. 2013YZ0002-3), the Scientific Foundation of Fuzhou Health Department (Grant no. 2013-S-wq18, and Grant no. 2013-S-wp1), the Mengchao Hepatobiliary Hospital of Fujian Medical University (Grant no. QDZJ-2014-004), the Science and Technology Bureau of Fuzhou City (Grand no. 2014-S-139-1), the Fujian Provincial Health and Family Planning Commission (Grant no. 2014-2-41), and the Research Development Special Fund of Indirectly Affiliated Hospital of Fujian Medical University (Grant no.FZS13004Y).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. 1
Hierarchical clustering of the differentially expressed LncRNAs and mRNAs in the primary HCC tissues from patients group T1, T2 and NT; T1 represents patients who had recurrence within one year after surgical resection and with outside liver metastasis; T2 represents patients who and recurrence after 2 years of surgical resection and without outside liver metastasis; NT represents normal liver tissues. (A) Differentially expressed LncRNAs between comparison of T1 and NT; (B) differentially expressed mRNAs between comparison of T2 and NT. Red color indicates the up-regulation and green color indicates the down regulation. (PDF 216 kb)
Fig. 2
The distribution of differentially expressed LncRNAs and mRNAs on each chromosome in the comparison between T1 and NT (A) and in the comparison between T2 and NT (B); T1 represents patients who had recurrence within one year after surgical resection and with outside liver metastasis; T2 represents patients who and recurrence after 2 years of surgical resection and without outside liver metastasis; NT represents normal liver tissues. The percentage of LncRNAs and mRNAs distributed on each chromosome, was calculated by the numbers of LncRNAs and mRNAs on each chromosome dividing the total numbers of differentially expressed LncRNAs and mRNAs; ch represents chromosome. (PDF 263 kb)
Fig. 3
The distribution of up- and down-regulated LncRNAs (A) and mRNAs (B) on each chromosome in the comparison between T1 and NT and the distribution of up- and down-regulated LncRNA (C) and mRNA (D) in the comparison between T2 and NT; T1 represents patients who had recurrence within one year after surgical resection and with outside liver metastasis; T2 represents patients who and recurrence after 2 years of surgical resection and without outside liver metastasis; NT represents normal liver tissues. (PDF 82 kb)
Rights and permissions
About this article
Cite this article
Gao, Y., Chen, G., Zeng, Y. et al. Invasion and metastasis-related long noncoding RNA expression profiles in hepatocellular carcinoma. Tumor Biol. 36, 7409–7422 (2015). https://doi.org/10.1007/s13277-015-3408-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-3408-0