Abstract
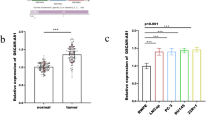
We aimed to evaluate the expression of microRNA-182 (miR-182) in triple-negative breast cancer (TNBC) tissues and the TNBC cell line MDA-MB-231 and to investigate the effects of mirR-182 on the cellular behavior of MDA-MB-231 and the expression of the target gene profilin 1 (PFN1), thus providing new methods and new strategies for the treatment of TNBC. Quantitative real-time PCR (qRT-PCR) was utilized to evaluate the expression of miR-182 in TNBC tissues, relatively normal tissues adjacent to TNBC and the TNBC cell line MDA-MB-231. Forty-eight hours after the MDA-MB-231 cells were transfected with the miR-182 inhibitor, qRT-PCR was utilized to detect the changes in miR-182 expression levels, and an MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay was utilized to determine the effects of miR-182 on cell viability. Flow cytometry was adopted to determine whether miR-182 affects the proliferation rates and apoptosis levels of the MDA-MB-231 cells. The transwell migration assay method was used to investigate the effects of miR-182 on the migration of the MDA-MB-231 cells. A luciferase reporter gene system was applied to validate that PFN1 was the target gene of miR-182. Western blot was used to measure the effects of miR-182 on the PFN1 protein expression levels in the cells. qRT-PCR results showed that compared with the relatively normal tissues adjacent to TNBC, miR-182 expression was significantly increased in the TNBC tissues and the MDA-MB-231 cells (p < 0.01). Compared with the control group, MDA-MB-231 cells transfected with the miR-182 inhibitor and incubated for 48 h showed significantly decreased miR-182 expression (p < 0.01). The results of an MTT assay showed that inhibition of miR-182 in MDA-MB-231 cells led to significantly reduced cell viability (p < 0.05). Flow cytometry analysis indicated that inhibition of miR-182 expression resulted in significantly decreased cell proliferation (p < 0.05) and significantly increased levels of apoptosis (p < 0.05). The results of a transwell migration assay showed that after inhibited of miR-182 expression, the number of cells passing through the transwell membranes was significantly decreased (p < 0.05). The results from a luciferase reporter gene system showed that compared with the control group, the relative luciferase activity of the group transfected with the miR-182 inhibitor was significantly increased (p < 0.05). Western blot analysis showed that compared with the control group, PFN1 protein expression levels were significantly increased in the MDA-MB-231 cells transfected with the miR-182 inhibitor and incubated for 48 h (p < 0.05). In conclusion, miR-182 is upregulated in TNBC tissues and cells. It promotes the proliferation and invasion of MDA-MB-231 cells and could negatively regulate PFN1 protein expression. Treatment strategies utilizing inhibition of miR-182 expression or overexpression of the PFN1 gene might benefit patients with TNBC.








Similar content being viewed by others
References
Ishikawa T, Shimizu D, Kito A, Ota I, Sasaki T, Tanabe M, et al. Breast cancer manifested by hematologic disorders. J Thorac Dis. 2012;4(6):650–4.
Arnedos M, Weigelt B, Reis-Filho JS. Anti-HER2 therapies: when more is more. Transl Cancer Res. 2012;1(1):49–54.
Fausto P, Sandro B. Benefit of tamoxifen in estrogen receptor positive DCIS of the breast. Gland Surg. 2012;1(1):3–4.
Rashid OM, Takabe K. The evolution of the role of surgery in the management of breast cancer lung metastasis. J Thorac Dis. 2012;4(4):420–4.
Nakhlis F, Golshan M. Bevacizumab: where do we go from here in breast cancer? Transl Cancer Res. 2012;1(1):55–6.
Hartman AR, Kaldate RR, Sailer LM, Painter L, Grier CE, Endsley RR, et al. Prevalence of BRCA mutations in an unselected population of triple-negative breast cancer. Cancer. 2012;118(11):2787–95.
Gholami S, Chen CH, Lou E, De Brot M, Fujisawa S, Chen NG, et al. Vaccinia Virus GLV-1h153 is effective in treating and preventing metastatic triple-negative breast cancer. Ann Surg. 2012;256(3):437–45.
Gelmon K, Dent R, Mackey JR, Laing K, McLeod D, Verma S. Targeting triple-negative breast cancer: optimising therapeutic outcomes. Ann Oncol. 2012;23(9):2223–34.
Patnaik SK, Mallick R, Yendamuri S. MicroRNAs and esophageal cancer. J Gastrointest Oncol. 2010;1(1):55–63.
Link A, Kupcinskas J, Wex T, Malfertheiner P. Macro-role of microRNA in gastric cancer. Dig Dis. 2012;30(3):255–67.
Santovito D, Mezzetti A, Cipollone F. MicroRNAs and atherosclerosis: new actors for an old movie. Nutr Metab Cardiovasc Dis. 2012;22(11):937–43.
Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13(4):271–82.
Ui-Tei K, Nishi K, Takahashi T, Nagasawa T. Thermodynamic control of small RNA-mediated gene silencing. Front Genet. 2012;3:101.
Yang JS, Maurin T, Lai EC. Functional parameters of dicer-independent microRNA biogenesis. RNA. 2012;18(5):945–57.
Chen J, Wang B. The roles of miRNA-143 in colon cancer and therapeutic implications. Transl Gastrointest Cancer. 2012;1(2):169–74.
Du L, Pertsemlidis A. microRNA regulation of cell viability and drug sensitivity in lung cancer. Expert Opin Biol Ther. 2012;12(9):1221–39.
Wall NR. Colorectal cancer screening using protected microRNAs. J Gastrointest Oncol. 2011;2(4):206–7.
Saus E, Soria V, Escaramis G, Vivarelli F, Crespo JM, Kagerbauer B, et al. Genetic variants and abnormal processing of pre-miR-182, a circadian clock modulator, in major depression patients with late insomnia. Hum Mol Genet. 2010;19(20):4017–25.
Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009;7(4):147–54.
Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci U S A. 2009;106(6):1814–9.
Santos A, Van Ree R. Profilins: mimickers of allergy or relevant allergens? Int Arch Allergy Immunol. 2011;155(3):191–204.
Gieni RS, Hendzel MJ. Actin dynamics and functions in the interphase nucleus: moving toward an understanding of nuclear polymeric actin. Biochem Cell Biol. 2009;87(1):283–306.
Rodriguez Faba O, Palou J. Predictive factors for recurrence progression and cancer specific survival in high-risk bladder cancer. Curr Opin Urol. 2012;22(5):415–20.
Qin XJ, Ling BX. Proteomic studies in breast cancer (Review). Oncol Lett. 2012;3(4):735–43.
Wei F, Xu J, Tang L, Shao J, Wang Y, Chen L, et al. p27(Kip1) V109G polymorphism and cancer risk: a systematic review and meta-analysis. Cancer Biother Radiopharm. 2012;27(10):665–71.
Wu Y, Sun WL, Feng JF. Antiangiogenic therapy in the management of breast cancer. Asia Pac J Clin Oncol. 2012. doi:10.1111/j.1743-7563.2012.01569.x.
Thorsen SB, Obad S, Jensen NF, Stenvang J, Kauppinen S. The therapeutic potential of microRNAs in cancer. Cancer J. 2012;18(3):275–84.
Cortez MA, Welsh JW, Calin GA. Circulating microRNAs as noninvasive biomarkers in breast cancer. Recent Results Cancer Res. 2012;195:151–61.
Vira D, Basak SK, Veena MS, Wang MB, Batra RK, Srivatsan ES. Cancer stem cells, microRNAs, and therapeutic strategies including natural products. Cancer Metastasis Rev. 2012;31(3–4):733–51.
Guan P, Yin Z, Li X, Wu W, Zhou B. Meta-analysis of human lung cancer microRNA expression profiling studies comparing cancer tissues with normal tissues. J Exp Clin Cancer Res. 2012;31(1):54.
Liu Z, Liu J, Segura MF, Shao C, Lee P, Gong Y, et al. MiR-182 overexpression in tumourigenesis of high-grade serous ovarian carcinoma. J Pathol. 2012;228(2):204–15.
Alshalalfa M, Bader GD, Goldenberg A, Morris Q, Alhajj R. Detecting microRNAs of high influence on protein functional interaction networks: a prostate cancer case study. BMC Syst Biol. 2012;6(1):112.
Weeraratne SD, Amani V, Teider N, Pierre-Francois J, Winter D, Kye MJ, et al. Pleiotropic effects of miR-183 96 182 converge to regulate cell survival, proliferation and migration in medulloblastoma. Acta Neuropathol. 2012;123(4):539–52.
Kong WQ, Bai R, Liu T, Cai CL, Liu M, Li X, et al. MicroRNA-182 targets cAMP-responsive element-binding protein 1 and suppresses cell growth in human gastric adenocarcinoma. FEBS J. 2012;279(7):1252–60.
Poell JB, van Haastert RJ, de Gunst T, Schultz IJ, Gommans WM, Verheul M, et al. A functional screen identifies specific microRNAs capable of inhibiting human melanoma cell viability. PLoS One. 2012;7(8):e43569.
Rawe VY, Payne C, Schatten G. Profilin and actin-related proteins regulate microfilament dynamics during early mammalian embryogenesis. Hum Reprod. 2006;21(5):1143–53.
Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol. 2002;4(8):626–31.
Wang D, Huang J, Hu Z. RNA helicase DDX5 regulates microRNA expression and contributes to cytoskeletal reorganization in basal breast cancer cells. Mol Cell Proteomics. 2012;11(2):M111 011932.
Zou L, Ding Z, Roy P. Profilin-1 overexpression inhibits proliferation of MDA-MB-231 breast cancer cells partly through p27kip1 upregulation. J Cell Physiol. 2010;223(3):623–9.
Acknowledgement
This study was supported by National Natural Science Foundation of China (No. 81150011).
Conflicts of interest
The authors have no commercial, proprietary, or financial interest in the products or companies described in this article.
Author information
Authors and Affiliations
Corresponding author
Additional information
Hailing Liu, Yan Wang, and Xin Li contributed equally to this article.
Rights and permissions
About this article
Cite this article
Liu, H., Wang, Y., Li, X. et al. Expression and regulatory function of miRNA-182 in triple-negative breast cancer cells through its targeting of profilin 1. Tumor Biol. 34, 1713–1722 (2013). https://doi.org/10.1007/s13277-013-0708-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-013-0708-0