Abstract
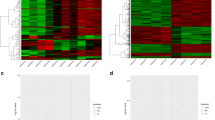
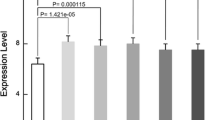
Spinal cord injury (SCI) remains to be the most devastating type of trauma for patients because of long lasting disability and limited response to the acute drug administration and efforts at rehabilitation. With the purpose to identify potential targets for SCI treatment and to gain more insights into the mechanisms of SCI, the microarray data of GSE2270, including 119 raphe magnus (RM) samples and 125 sensorimotor cortex (SMTC) samples, was downloaded from the Gene Expression Omnibus database. Differentially expressed genes (DEGs) were screened in RM group and SMTC group compared with their corresponding controls, respectively. A protein–protein interaction (PPI) network was constructed based on the common DEGs identified in both RM group and SMTC group. Gene ontology (GO) and pathway enrichment analyses of the overlapping DEGs were performed. Furthermore, the common DEGs enriched in each pathway were analyzed to identify significant regulatory elements. Totally, 173 overlapping DEGs (130 up-regulated and 43 down-regulated) were identified in both RM and SMTC samples. These overlapping DEGs were enriched in different GO terms. Pathway enrichment analysis revealed that DEGs were mainly related to inflammation and immunity. CD68 molecule (CD68) was a hub protein in the PPI network. Moreover, the regulatory network showed that ras-related C3 botulinum toxin substrate 2 (RAC2), CD44 molecule (CD44), and actin related protein 2/3 complex (ARPC1B) were hub genes. RAC2, CD44, and ARPC1B may be significantly involved in the pathogenesis of SCI by participating significant pathways such as extracellular matrix-receptor signaling pathway and Toll-like receptor signaling pathway.



Similar content being viewed by others
References
Ackery A, Tator C, Krassioukov A (2004) A global perspective on spinal cord injury epidemiology. J Neurotrauma 21:1355–1370
Adair-Kirk TL, Senior RM (2008) Fragments of extracellular matrix as mediators of inflammation. Int J Biochem Cell Biol 40:1101–1110
Almad A, Sahinkaya FR, McTigue DM (2011) Oligodendrocyte fate after spinal cord injury. Neurotherapeutics 8:262–273
Basso DM (2000) Neuroanatomical substrates of functional recovery after experimental spinal cord injury: implications of basic science research for human spinal cord injury. Phys Ther 80:808–817
Bland JM, Altman DG (1995) Multiple significance tests: the Bonferroni method. BMJ 310:170
Boekhoff TM, Ensinger EM, Carlson R, Bock P, Baumgartner W, Rohn K, Tipold A, Stein VM (2012) Microglial contribution to secondary injury evaluated in a large animal model of human spinal cord trauma. J Neurotrauma 29:1000–1011
Boyer L, Magoc L, Dejardin S, Cappillino M, Paquette N, Hinault C, Charriere GM, Ip W, Fracchia S, Hennessy E (2011) Pathogen-derived effectors trigger protective immunity via activation of the Rac2 enzyme and the IMD or Rip kinase signaling pathway. Immunity 35:536–549
Budh CN, Osteraker AL (2007) Life satisfaction in individuals with a spinal cord injury and pain. Clin Rehabil 21:89–96
Buss A, Pech K, Kakulas BA, Martin D, Schoenen J, Noth J, Brook GA (2007) Matrix metalloproteinases and their inhibitors in human traumatic spinal cord injury. BMC Neurol 7:17
Byrnes KR, Faden AI (2007) Role of cell cycle proteins in CNS injury. Neurochem Res 32:1799–1807
Byrnes KR, Garay J, Di Giovanni S, De Biase A, Knoblach SM, Hoffman EP, Movsesyan V, Faden AI (2006) Expression of two temporally distinct microglia-related gene clusters after spinal cord injury. Glia 53:420–433
Byrnes KR, Stoica BA, Fricke S, Di Giovanni S, Faden AI (2007) Cell cycle activation contributes to post-mitotic cell death and secondary damage after spinal cord injury. Brain 130:2977–2992
Citron BA, Arnold PM, Sebastian C, Qin F, Malladi S, Ameenuddin S, Landis ME, Festoff BW (2000) Rapid upregulation of caspase-3 in rat spinal cord after injury: mRNA, protein, and cellular localization correlates with apoptotic cell death. Exp Neurol 166:213–226
Cortez R, Levi AD (2007) Acute spinal cord injury. Curr Treat Options Neurol 9:115–125
Cruse J, Lewis R, Dilioglou S, Roe D, Wallace W, Chen R (1999) Review of immune function, healing of pressure ulcers, and nutritional status in patients with spinal cord injury. J Spinal Cord Med 23:129–135
Cui X, Churchill GA (2003) Statistical tests for differential expression in cDNA microarray experiments. Genome Biol 4:210
De Biase A, Knoblach SM, Di Giovanni S, Fan C, Molon A, Hoffman EP, Faden AI (2005) Gene expression profiling of experimental traumatic spinal cord injury as a function of distance from impact site and injury severity. Physiol Genomics 22:368–381
Desborough J (2000) The stress response to trauma and surgery. Br J Anaesth 85:109–117
Di Giovanni S, Knoblach SM, Brandoli C, Aden SA, Hoffman EP, Faden AI (2003) Gene profiling in spinal cord injury shows role of cell cycle in neuronal death. Ann Neurol 53:454–468
Diboun I, Wernisch L, Orengo CA, Koltzenburg M (2006) Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics 7:252
Donnelly DJ, Popovich PG (2008) Inflammation and its role in neuroprotection, axonal regeneration and functional recovery after spinal cord injury. Exp Neurol 209:378–388
Efron B, Tibshirani R (2002) Empirical Bayes methods and false discovery rates for microarrays. Genet Epidemiol 23:70–86
Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20:307–315
Gomez-Cambronero J (2014) Phosphatidic acid, phospholipase D and tumorigenesis. Adv Biol Regul 54:197–206
Gotea V, Ovcharenko I (2008) DiRE: identifying distant regulatory elements of co-expressed genes. Nucleic Acids Res 36:W133–W139
Grossman SD, Wolfe BB, Yasuda RP, Wrathall JR (2000) Changes in NMDA receptor subunit expression in response to contusive spinal cord injury. J Neurochem 75:174–184
Hauben E, Schwartz M (2003) Therapeutic vaccination for spinal cord injury: helping the body to cure itself. Trends Pharmacol Sci 24:7–12
Hayashi M, Ueyama T, Nemoto K, Tamaki T, Senba E (2000) Sequential mRNA expression for immediate early genes, cytokines, and neurotrophins in spinal cord injury. J Neurotrauma 17:203–218
Houle JD, Tessler A (2003) Repair of chronic spinal cord injury. Exp Neurol 182:247–260
Ietta F, Wu Y, Winter J, Xu J, Wang J, Post M, Caniggia I (2006) Dynamic HIF1A regulation during human placental development. Biol Reprod 75:112–121
Ihaka R, Gentleman R (1996) R: a language for data analysis and graphics. J Comput Graph Stat 5:299–314
Joshi S, Singh AR, Zulcic M, Bao L, Messer K, Ideker T, Dutkowski J, Durden DL (2014) Rac2 controls tumor growth, metastasis and M1-M2 macrophage differentiation in vivo. PLoS One 9:e95893
Kaisho T, Akira S (2006) Toll-like receptor function and signaling. J Allergy Clin Immunol 117:979–987
Kigerl KA, Lai W, Rivest S, Hart RP, Satoskar AR, Popovich PG (2007) Toll-like receptor (TLR)-2 and TLR-4 regulate inflammation, gliosis, and myelin sparing after spinal cord injury. J Neurochem 102:37–50
Lee JY, Chung H, Yoo YS, Oh YJ, Oh TH, Park S, Yune TY (2010) Inhibition of apoptotic cell death by ghrelin improves functional recovery after spinal cord injury. Endocrinology 151:3815–3826
Liebl DJ, Huang W, Young W, Parada LF (2001) Regulation of Trk receptors following contusion of the rat spinal cord. Exp Neurol 167:15–26
Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21:3448–3449
May R (2001) The Arp2/3 complex: a central regulator of the actin cytoskeleton. Cell Mol Life Sci 58:1607–1626
McDonald JW, Sadowsky C (2002) Spinal-cord injury. Lancet 359:417–425
Molli PR, Li D-Q, Bagheri-Yarmand R, Pakala SB, Katayama H, Sen S, Iyer J, Chernoff J, Tsai M-Y, Nair SS (2010) Arpc1b, a centrosomal protein, is both an activator and substrate of Aurora A. J Cell Biol 190:101–114
Oyinbo CA (2011) Secondary injury mechanisms in traumatic spinal cord injury: a nugget of this multiply cascade. Acta Neurobiol Exp (Wars) 71:281–299
Papa S, Ferrari R, De Paola M, Rossi F, Mariani A, Caron I, Sammali E, Peviani M, Dell’Oro V, Colombo C (2014) Polymeric nanoparticle system to target activated microglia/macrophages in spinal cord injury. J Control Release 174:15–26
Parker HS, Leek JT, Favorov AV, Considine M, Xia X, Chavan S, Chung CH, Fertig EJ (2014) Preserving biological heterogeneity with a permuted surrogate variable analysis for genomics batch correction. Bioinformatics 30:2757–2763
Plowden J, Renshaw-Hoelscher M, Engleman C, Katz J, Sambhara S (2004) Innate immunity in aging: impact on macrophage function. Aging Cell 3:161–167
Ponta H, Sherman L, Herrlich PA (2003) CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol 4:33–45
Profyris C, Cheema SS, Zang D, Azari MF, Boyle K, Petratos S (2004) Degenerative and regenerative mechanisms governing spinal cord injury. Neurobiol Dis 15:415–436
Roberts AW, Kim C, Zhen L, Lowe JB, Kapur R, Petryniak B, Spaetti A, Pollock JD, Borneo JB, Bradford GB (1999) Deficiency of the hematopoietic cell-specific Rho family GTPase Rac2 is characterized by abnormalities in neutrophil function and host defense. Immunity 10:183–196
Satake K, Matsuyama Y, Kamiya M, Kawakami H, Iwata H, Adachi K, Kiuchi K (2000) Up-regulation of glial cell line-derived neurotrophic factor (GDNF) following traumatic spinal cord injury. NeuroReport 11:3877–3881
Sherman LS, Rizvi TA, Karyala S, Ratner N (2000) CD44 enhances neuregulin signaling by Schwann cells. J Cell Biol 150:1071–1084
Sherman BT, Huang DW, Tan Q, Guo Y, Bour S, Liu D, Stephens R, Baseler MW, Lane HC, Lempicki RA (2007) DAVID Knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics 8:426
Skinner M (2010) Cell cycle: ARPC1B—a regulator of regulators. Nat Rev Mol Cell Biol 11:542
Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27:431–432
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452
Tang Y, Li M, Wang J, Pan Y, Wu F-X (2015) CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. BioSystems 127:67–72
Xia X, Qu B, Ma Y, L Yang, H Huang, J Cheng, Yang T, Kong B, E Liu, Zhao K (2014) Analyzing time-series microarray data reveals key genes in spinal cord injury. Mol Biol Rep 41:6827–6835
Xiaowei H, Ninghui Z, Wei X, Yiping T, Linfeng X (2005) The experimental study of hypoxia-inducible factor-1α and its target genes in spinal cord injury. Spinal Cord 44:35–43
Yang N, Farrell A, Niedelman W, Melo M, Lu D, Julien L, Marth GT, Gubbels M-J, Saeij JP (2013) Genetic basis for phenotypic differences between different Toxoplasma gondii type I strains. BMC Genomics 14:467
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declared that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Peng, D., Chen, M., Zuo, G. et al. Analysis of the potential pathways and target genes in spinal cord injury using bioinformatics methods. Genes Genom 38, 619–628 (2016). https://doi.org/10.1007/s13258-016-0385-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-016-0385-7