Abstract
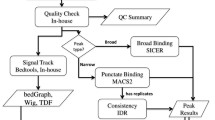
Although there are many applications available for the analysis of chromatin immunoprecipitation with massively parallel DNA sequencing (ChIP-seq), users need some knowledge about the installation, alignment, and peak calling procedures prior to the analysis. Here, we present an easy-to-use application for ChIP-seq analysis called AutoChIP. With AutoChIP, installation of necessary programs, alignment of unmapped reads to a reference genome, and identification of genome-wide binding sites can be done in a single step with a large set of ChIP-seq data. Evaluation of the cocktail algorithm implemented in AutoChIP showed that it outperformed a single ChIP-seq tool in terms of the ratio of motif occurrences and the average height of normalized read density over the identified peaks. In addition, annotation of the identified peaks with the known gene and repeat elements information provides a comprehensive picture of the genome-wide binding sites of given proteins. Overall, AutoChIP provides a comprehensive platform to analyze a large set of ChIP-seq data in one step.




Similar content being viewed by others
References
Adomas AB, Grimm SA, Malone C, Takaku M, Sims JK, Wade PA (2014) Breast tumor specific mutation in GATA3 affects physiological mechanisms regulating transcription factor turnover. BMC Cancer 14:278
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M et al (2013) NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res 41:D991–D995
Feng X, Grossman R, Stein L (2011) PeakRanger: a cloud-enabled peak caller for ChIP-seq data. BMC Bioinformatics 12:139
Feuermann Y, Kang K, Gavrilova O, Haetscher N, Jang SJ, Yoo KH, Jiang C, Gonzalez FJ, Robinson GW, Hennighausen L (2013) MiR-193b and miR-365-1 are not required for the development and function of brown fat in the mouse. RNA Biol 10:1807–1814
Gonsky R, Deem RL, Bream J, Young HA, Targan SR (2004) Enhancer role of STAT5 in CD2 activation of IFN- gene expression. J Immunol 173:6241–6247
Grant CE, Bailey TL, Noble WS (2011) FIMO: scanning for occurrences of a given motif. Bioinformatics 27:1017–1018
Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38:576–589
Henry VJ, Bandrowski AE, Pepin AS, Gonzalez BJ, Desfeux A (2014) OMICtools: an informative directory for multi-omic data analysis. Database (Oxford) 2014. doi:10.1093/database/bau069
Kang K, Robinson GW, Hennighausen L (2013) Comprehensive meta-analysis of signal transducers and activators of transcription (STAT) genomic binding patterns discerns cell-specific cis-regulatory modules. BMC Genom 14:4
Kang K, Yamaji D, Yoo KH, Robinson GW, Hennighausen L (2014) Mammary-specific gene activation is defined by progressive recruitment of STAT5 during pregnancy and the establishment of H3K4me3 marks. Mol Cell Biol 34:464–473
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Liao Y, Smyth GK, Shi W (2013) The subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41:e108. doi:10.1093/nar/gkt214
Machanick P, Bailey TL (2011) MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27:1696–1697
Malone BM, Tan F, Bridges SM, Peng Z (2011) Comparison of four ChIP-Seq analytical algorithms using rice endosperm H3K27 trimethylation profiling data. PLoS One 6:e25260. doi:10.1371/journal.pone.0025260
Mouse EC, Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R et al (2012) An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol 13:418
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842
Ranganath S, Ouyang W, Bhattarcharya D, Sha WC, Grupe A, Peltz G, Murphy KM (1998) GATA-3-dependent enhancer activity in IL-4 gene regulation. J Immunol 161:3822–3826
Yamaji D, Kang K, Robinson GW, Hennighausen L (2013) Sequential activation of genetic programs in mouse mammary epithelium during pregnancy depends on STAT5A/B concentration. Nucleic Acids Res 41:1622–1636
Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W et al (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137
Acknowledgments
We thank members of the Kang laboratory for valuable comments.
Conflict of interest
The authors state that there are no conflicts of interest.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kim, T., Lee, W., Han, K. et al. An automated analysis pipeline for a large set of ChIP-seq data: AutoChIP. Genes Genom 37, 305–311 (2015). https://doi.org/10.1007/s13258-014-0260-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-014-0260-3