Abstract
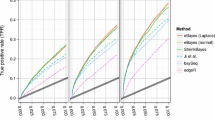
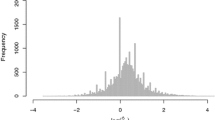
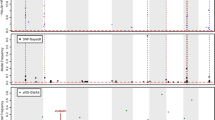
The performance of inbred and hybrid genotypes is of interest in plant breeding and genetics. High-throughput sequencing of RNA (RNA-seq) has proven to be a useful tool in the study of the molecular genetic responses of inbreds and hybrids to environmental stresses. Commonly used experimental designs and sequencing methods lead to complex data structures that require careful attention in data analysis. We demonstrate an analysis of RNA-seq data from a split-plot design involving drought stress applied to two inbred genotypes and two hybrids formed by crosses between the inbreds. Our generalized linear modeling strategy incorporates random effects for whole-plot experimental units and uses negative binomial distributions to allow for overdispersion in count responses for split-plot experimental units. Variations in gene length and base content, as well as differences in sequencing intensity across experimental units, are also accounted for. Hierarchical modeling with thoughtful parameterization and prior specification allows for borrowing of information across genes to improve estimation of dispersion parameters, genotype effects, treatment effects, and interaction effects of primary interest.






Similar content being viewed by others
References
Anders, S., and Huber, W. (2010), “Differential expression analysis for sequence count data,” Genome Biol, 11(10), R106.
Benjamini, Y., and Speed, T. P. (2012), “Summarizing and correcting the GC content bias in high-throughput sequencing,” Nucleic Acids Research, p. gks001.
Dillies, M.-A., Rau, A., Aubert, J., Hennequet-Antier, C., Jeanmougin, M., Servant, N., Keime, C., Marot, G., Castel, D., Estelle, J. et al. (2013), “A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis,” Briefings in Bioinformatics, 14(6), 671–683.
Hardcastle, T. J., and Kelly, K. A. (2010), “baySeq: empirical Bayesian methods for identifying differential expression in sequence count data,” BMC Bioinformatics, 11(1), 422.
Law, C. W., Chen, Y., Shi, W., and Smyth, G. K. (2014), “Voom: precision weights unlock linear model analysis tools for RNA-seq read counts,” Genome Biol, 15(2), R29.
Lewin, A., Bochkina, N., and Richardson, S. (2007), “Fully Bayesian mixture model for differential gene expression: simulations and model checks,” Statistical Applications in Genetics and Molecular Biology, 6(1).
Li, J., and Tibshirani, R. (2013), “Finding consistent patterns: a nonparametric approach for identifying differential expression in RNA-seq data,” Statistical Methods in Medical Research, 22(5), 519–536.
Lorenz, D. J., Gill, R. S., Mitra, R., and Datta, S. (2014), “Using RNA-seq Data to Detect Differentially Expressed Genes,” in Statistical Analysis of Next Generation Sequencing Data Springer, pp. 25–49.
Love, M. I., Huber, W., and Anders, S. (2014), “Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2,” Genome Biol, 15(12), 550.
Lund, S. P., Nettleton, D. et al. (2012), “The importance of distinct modeling strategies for gene and gene-specific treatment effects in hierarchical models for microarray data,” The Annals of Applied Statistics, 6(3), 1118–1133.
McCarthy, D. J., Chen, Y., and Smyth, G. K. (2012), “Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation,” Nucleic Acids Research, 40(10), 4288–4297.
Nettleton, D. (2014), “Design of RNA Sequencing Experiments,” in Statistical Analysis of Next Generation Sequencing Data Springer, pp. 93–113.
Oshlack, A., Wakefield, M. J. et al. (2009), “Transcript length bias in RNA-seq data confounds systems biology,” Biol Direct, 4(1), 14.
Riebler, A., Robinson, M. D., and van de Wiel, M. A. (2014), “Analysis of Next Generation Sequencing Data Using Integrated Nested Laplace Approximation (INLA),” in Statistical Analysis of Next Generation Sequencing Data Springer, pp. 75–91.
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010), “edgeR: a Bioconductor package for differential expression analysis of digital gene expression data,” Bioinformatics, 26(1), 139–140.
Robinson, M. D., Oshlack, A. et al. (2010), “A scaling normalization method for differential expression analysis of RNA-seq data,” Genome Biol, 11(3), R25.
Rue, H., Martino, S., and Chopin, N. (2009), “Approximate Bayesian inference for latent Gaussian models by using integrated nested Laplace approximations,” Journal of the Royal Statistical Society: Series B (Statistical Methodology), 71(2), 319–392.
Smyth, G. K. (2005), “Limma: linear models for microarray data,” in Bioinformatics and computational biology solutions using R and Bioconductor Springer, pp. 397–420.
van de Wiel, M. A., Leday, G. G., Pardo, L., Rue, H., Van Der Vaart, A. W., and Van Wieringen, W. N. (2013), “Bayesian analysis of RNA sequencing data by estimating multiple shrinkage priors,” Biostatistics, 14(1), 113–128.
Ventrucci, M., Scott, E. M., and Cocchi, D. (2011), “Multiple testing on standardized mortality ratios: a Bayesian hierarchical model for FDR estimation,” Biostatistics, 12(1), 51–67.
Acknowledgments
Research reported in this chapter was supported by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health and the joint National Science Foundation/NIGMS Mathematical Biology Program under award number R01GM109458. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health or the National Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lithio, A., Nettleton, D. Hierarchical Modeling and Differential Expression Analysis for RNA-seq Experiments with Inbred and Hybrid Genotypes. JABES 20, 598–613 (2015). https://doi.org/10.1007/s13253-015-0232-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13253-015-0232-3