Abstract
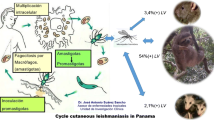
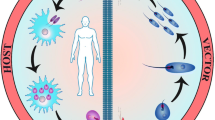
Leishmaniasis, one of the neglected tropical diseases is serious health concern globally. The disease is caused by protozoan parasites belonging to genus Leishmania. The main forms of disease are Cutaneous Leishmaniasis (CL), Mucocutaneous Leishmaniasis and Visceral Leishmaniasis (VL). VL or Kala-azar is the most severe form and 90% of global VL cases occur in India, Bangladesh, Nepal, Sudan, Ethiopia and Brazil, while most cases (70–75%) of CL occur in Afghanistan, Brazil, Iran, Ethiopia, Costa Rica and Peru etc. They are spread by the bites of female sand flies of the genus Phlebotomus in the Old World and of the genus Lutzomyia in the New World. It is essential to determine whether genetic variability of the parasites is associated with the different clinical manifestations and drug resistance of Leishmania sp. Various molecular biological methods have been standardized to study the genomes of the parasites in order to understand the parasites better. Most updated high-throughput approaches are whole genome sequencing, comparative genomics, transcriptomics and proteomics. The present review gives an overview of the advancement in the field of the Leishmania genome analysis which would help workers in the field to understand the problem of emergence of drug resistance, current epidemiological status, host parasite interaction and designing the drugs.
Similar content being viewed by others
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402.
Alvar J, Velez ID, Bern C, Herrero M, Desjeux P, Cano J, Jannin J, den Boer M. Leishmaniasis worldwide and global estimates of its incidence. PLoS ONE. 2012;7:e35671.
Baldauf SL. The deep roots of eukaryotes. Science. 2003;300:1703–6.
Bañuls AL, Hide M, Prugnolle F. Leishmania and the Leishmaniases: a parasite genetic update and advances in taxonomy, epidemiology and pathogenicity in humans. Adv Parasitol. 2007;64:1–109.
Bao Y, Weiss LM, Braunstein VL, Huang H. Role of protein kinase A in Trypanosoma cruzi. Infect Immun. 2008;76:4757–63.
Bard E. Molecular biology of Leishmania. Biochem Cell Biol. 1989;67:516–24.
Bayona JC, Nakayasu ES, Laverrière M, Aguilar C, Sobreira TJ, Choi H, et al. SUMOylation pathway in Trypanosoma cruzi: functional characterization and proteomic analysis of target proteins. Mol Cell Proteomics. 2011;10(M110):007369.
Bettini S, Maroli M, Gradoni L. Leishmaniasis in Tuscany (Italy): (IV) An analysis of all recorded human cases. Trans R Soc Trop Med Hyg. 1981;75:338–44.
Beverley SM. Characterization of the ‘unusual’ mobility of large circular DNAs in pulsed field-gradient electrophoresis. Nucleic Acids Res. 1988;16:925–39.
Beverley SM. Gene amplification in Leishmania. Annual Rev Microbiol. 1991;45:417–44.
Blaineau C, Bastien P, Rioux JA, Roizes G, Pagès M. Long-range restriction maps of size-variable homologous chromosomes in Leishmania infantum. Mol Biochem Parasitol. 1991;46:292–302.
Brinkworth RI, Breinl RA, Kobe B. Structural basis and prediction of substrate specificity in protein serine/threonine kinases. Proc Natl Acad Sci USA. 2003;100:74–9.
Brisse S, Dujardin JC, Tibayrenc M. Identification of six Trypanosoma cruzi lineages by sequence-characterised amplified region markers. Mol Biochem Parasitol. 2000;111:95–105.
Britto C, Ravel C, Bastien P, Blaineau C, Page’s M, Dedet JP, et al. Conserved linkage groups associated with large-scale chromosomal rearrangements between Old World and New World Leishmania genomes. Gene. 1998;222:107–17.
Croft SL, Sundar S, Fairlamb AH. Drug resistance in Leishmaniasis. Clin Microbiol Rev. 2006;19:111–26.
de Paiva RM, Grazielle-Silva V, Cardoso MS, Nakagaki BN, Mendonça-Neto RP, Canavaci AM, et al. Amastin Knockdown in Leishmania braziliensis affects parasite-macrophage interaction and results in impaired viability of intracellular amastigotes. PLoS Pathog. 2015;11:e1005296.
Desjeux P. WHO/CID internal document, the leishmaniasis, meeting of interested parties on management and financing of the control of tropical diseases other than malaria. Geneva, CTD/MIP/WP/93.8; 1993.
Desjeux P, Alvar J. Leishmania/HIV co-infections: epidemiology in Europe. Ann Trop Med Parasitol. 2003;97(suppl):3–15.
Donelson JE, Zeng W. A comparison of trans-RNA splicing in trypanosomes and nematodes. Parasitol Today. 1990;6(10):327–34.
Downing T, Imamura H, Decuypere S, Clark TG, Coombs GH, Cotton JA, et al. Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res. 2011;21:2143–56.
El-Sayed NM, Myler PJ, Blandin G, Berriman M, Crabtree J, Aggarwal G, et al. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309:404–9.
El Tai NO, Osman OF, El Fari M, Presber W, Schonian G. Genetic heterogeneity of ribosomal internal transcribed spacer in clinical samples of Leishmania donovani spotted on filter paper as revealed by single-strand conformation polymorphisms and sequencing. Trans R Soc Trop Med Hyg. 2000;94:575–9.
Enright MC, Spratt BG. Multilocus sequence typing. Trends Microbiol. 1999;7:482–7.
Fantoni A, Dare AO, Tschudi C. RNA polymerase III-mediated transcription of the trypanosome U2 small nuclear RNA gene is controlled by both intragenic and extragenic regulatory elements. Mol Cell Biol. 1994;14:2021–8.
Fernandez-Moya SM, Estevez AM. Posttranscriptional control and the role of RNA-binding proteins in gene regulation in trypanosomatid protozoan parasites. Wiley Interdiscip Rev RNA. 2010;1:34–46.
Gannavaram S, Sharma P, Duncan RC, Salotra P, Nakhasi HL. Mitochondrial associated ubiquitin fold modifier-1 mediated protein conjugation in Leishmania donovani. PLoS ONE. 2011;6:e16156.
Hartley MA, Ronet C, Zangger H, Beverley SM, Fasel N. Leishmania RNA virus: when the host pays the toll. Front Cell Infect Microbiol. 2012;2:99.
Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12.
Hofmann K, Bucher P, Falquet L, Bairoch A. The PROSITE database, its status in 1999. Nucleic Acids Res. 1999;27:215–9.
Imamura H, Downing T, Van den Broeck F, Sanders MJ, Rijal S, Sundar S, et al. Evolutionary genomics of epidemic visceral leishmaniasis in the Indian subcontinent. ELife. 2016;5:e12613.
Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–42.
Kapler GM, Beverley SM. Transcriptional mapping of the amplified region encoding the dihydrofolate reductase-thymidylate synthase of Leishmania major reveals a high density of transcripts, including overlapping and antisense RNAs. Mol Cell Biol. 1989;9:3959–72.
Khanra S, Bandopadhyay SK, Chakraborty P, Datta S, Mondal D, Chatterjee M, et al. Characterization of the recent clinical isolates of Indian Kala-azar patients by RAPD-PCR method. J Parasit Dis. 2011;35:116–22.
Khanra S, Datta S, Mondal D, Saha P, Bandopadhyay SK, Roy S, et al. RFLPs of ITS, ITS1 and hsp70 amplicons and sequencing of ITS1 of recent clinical isolates of Kala-azar from India and Bangladesh confirms the association of L. tropica with the disease. Acta Trop. 2012;124:229–34.
Khanra S, Manna M. Molecular tools for Leishmania sp. identification and characterization. Perspect Cytol Genet. 2011;15:292–300.
Khanra S, Sarraf NR, Das S, Das AK, Roy S, Manna M. Genetic markers for antimony resistant clinical isolates differentiation from Indian Kala-azar. Acta Trop. 2016;164:177–84.
Kramer S. Developmental regulation of gene expression in the absence of transcriptional control: the case of kinetoplastids. Mol Biochem Parasitol. 2012;181:61–72.
Kumar A, Boggula VR, Misra P, Sundar S, Shasany AK, Dube A. Amplified fragment length polymorphism (AFLP) analysis is useful for distinguishing Leishmania species of visceral and cutaneous forms. Acta Trop. 2010;113:202–6.
Kumar A, Boggula VR, Sundar S, Shasany AK, Dube A. Identification of genetic markers in sodium antimony gluconate (SAG) sensitive and resistant Indian clinical isolates of Leishmania donovani through amplified fragment length polymorphism (AFLP). Acta Trop. 2009;110:80–5.
LeBowitz JH, Smith HQ, Rusche L, Beverley SM. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev. 1993;7:996–1007.
León AM, Shaw JJ, Tapia FJ. A guide for the cutaneous leishmaniasis connoisseur. In: Tapia FJD, Dittmar GC, Sànchez MA, editors. Molecular and immune mechanisms in the pathogenesis of Cutaneous leishmaniasis. Austin: Academic Press and R.G. Landes Company; 1996. p. 1–23.
Lewin S, Schonian G, El Tai N, Oskam L, Bastien P, Presber W. Strain typing in Leishmania donovani by using sequence-confirmed amplified region analysis. Int J Parasitol. 2002;32:1267–76.
Liu L, Li Y, Li S, Hu N, He Y, Pong R, et al. Comparison of next-generation sequencing systems. J Biomed Biotechnol. 2012;2012:251364.
Lye LF, Owens K, Shi H, Murta SMF, Vieira AC, Turco SJ, et al. Retention and loss of RNA interference pathways in Trypanosomatid protozoans. PLoS Pathog. 2010;6:e1001161.
Maity AK, Goswami A, Saha P. Identification of substrates of an S-phase cell cycle kinase from Leishmania donovani. FEBS Lett. 2011;585:2635–9.
Mallik KK, Basu D. Recent advances in the management of Indian Kala-azar. In: Bhaduri AN, Basu MK, Sen AK, Kumar S, editors. Current trends in Leishmania research. New Delhi: Publication of Information Directorate (CSIR); 1993. p. 263.
Manna M, Khanra S. Molecular identification of an old clinical isolate of Indian Kala-azar. J Parasit Dis. 2016; doi:10.1007/s12639-016-0799-9.
Manna M, Majumder H, Sundar S, Bhaduri A. Molecular characterizations of the clinical isolates of Kala-azar patients of India suggest Leishmania donovani is the only causal agent of Indian Kala-azar. Med Sci Monit. 2005;11:220–7.
Martinez-Calvillo S, Nguyen D, Stuart K, Myler P. Transcription initiation and termination on Leishmania major chromosome 3. Eukaryot Cell. 2004;3:506–17.
Masiga DK, Turner CM. Amplified (restriction) fragment length polymorphism (AFLP) analysis. Methods Mol Biol. 2004;270:173–86.
Moreira D, Lopez-Garcia P, Vickerman K. An updated view of kinetoplastid phylogeny using environmental sequences and a closer outgroup: proposal for a new classification of the class Kinetoplastea. Int J Syst Evol Microbiol. 2004;54:1861–75.
Mukhopadhyay R, Mukherjee S, Mukherjee B, Naskar K, Mondal D, Decuypere S, et al. Characterization of antimony-resistant Leishmania donovani isolates: biochemical and biophysical studies and interaction with host cells. Int J Parasitol. 2011;41:1311–21.
Murray HW, Berman JD, Davies CR, Saravia NG. Advances in leishmaniasis. Lancet. 2005;66:1561–77.
Myler PJ, Audleman L, DeVos T, Hixson G, Kiser P, Lemley C, et al. Leishmania major Friedlin chromosome 1 has an unusual distribution of protein-coding genes. Proc Natl Acad Sci USA. 1999;96:2902–6.
Naik SR, Rao PN, Datta DV, Mehta SK, Mahajan RC, Mehta S, et al. Kala-azar in north-western India: a study of 24 patients. Trans R Soc Trop Med Hyg. 1979;73:61–5.
Nowrousian M. Next-generation sequencing techniques for eukaryotic microorganisms: sequencing-based solutions to biological problems. Eukaryot Cell. 2010;9:1300–10.
Otranto D, Dantas-Torres F. The prevention of canine leishmaniasis and its impact on public health. Trends Parasitol. 2013;29:339–45.
Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, Quail MA, et al. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet. 2007;39:839–47.
Rastrojo A, Carrasco-Ramiro F, Martín D, Crespillo A, Reguera RM, Aguado B, et al. The transcriptome of Leishmania major in the axenic promastigote stage: transcript annotation and relative expression levels by RNA-seq. BMC Genom. 2013;14:223.
Ravel C, Dubessay P, Britto C, Blaineau C, Bastien P, Pagès M. High conservation of the fine-scale organisation of chromosome 5 between two pathogenic Leishmania species. Nucleic Acids Res. 1999;27:2473–7.
Real F, Vidal RO, Carazzolle MF, Mondego JM, Costa GG, Herai RH, et al. The genome sequence of Leishmania (Leishmania) amazonensis: functional annotation and extended analysis of gene models. DNA Res. 2013;20:567–81.
Requena JM. Lights and shadows on gene organization and regulation of gene expression in Leishmania. Front Biosci. 2011;17:2069–85.
Rogers MB, Hilley JD, Dickens NJ, Wilkes J, Bates PA, Depledge DP, et al. Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res. 2011;21:2129–42.
Rojas-Sánchez S, Figueroa-Angulo E, Moreno-Campos R, Florencio-Martínez LE, Manning-Cela RG, Martínez-Calvillo S. Transcription of Leishmania major U2 small nuclear RNA gene is directed by extragenic sequences located within a tRNA-like and a tRNA-Ala gene. Parasit Vectors. 2016;9:401.
Rosenzweig D, Smith D, Myler PJ, Olafson RW, Zilberstein D. Post-translational modification of cellular proteins during Leishmania donovani differentiation. Proteomics. 2008;8:1843–50.
Sacks DL, Kenney RT, Kreutzer RD, Jaffe CL, Gupta AK, Sharma MC, et al. Indian Kala-azar caused by L. tropica. Lancet. 1995;345:959–61.
Simpson L. The mitochondrial genome of kinetoplastid protozoa: genomic organization, transcription, replication and evolution. Ann Rev Microbiol. 1987;41:363–82.
Sinha S, Sundaram S. An analysis of phosphorylation sites in protein kinases from Leishmania. Bioinformation. 2016;12:249–53.
Srivastava P, Prajapati VK, Vanaerschot M, Van der Auwera G, Dujardin JC, Sundar S. Detection of Leptomonas sp. parasites in clinical isolates of Kala-azar patients from India. Infect Genet Evol. 2010;10:1145–50.
Srivastava P, Singh T, Sundar S. Genetic heterogeneity in clinical isolates of Leishmania donovani from India. J Clin Microbiol. 2011;49:3687–90.
Sturm NR, Simpson L. Kinetoplast DNA minicircles encode guide RNAs for editing of cytochrome oxidase subunit III mRNA. Cell. 1990;61:879–84.
Taylor JW, Geiser DM, Burt A, Koufopanou V. The evolutionary biology and population genetics underlying fungal strain typing. Clin Microbiol Rev. 1999;12:126–46.
Thakur CP. Diminishing effectiveness of currently used drugs in treatment of Kala-azar and Ampphotericine B in antimony and pentamidine resistant Kala-azar. In: Bhaduri AN, Basu MK, Sen AK, Kumar S, editors. Current trends in Leishmania research. New Delhi: Publication and Information Directorate (CSIR); 1993. p. 254.
Thomas S, Green A, Sturm NR, Campbell DA, Myler PJ. Histone acetylations mark origins of polycistronic transcription in Leishmania major. BMC Genom. 2009;10:152.
van Luenen HGAM, Farris C, Jan S, Genest PA, Tripathi P, Velds A, et al. Glucosylated hydroxymethyluracil, DNA base J, prevents transcriptional readthrough in Leishmania. Cell. 2012;150:909–21.
Velez-Ramirez DE, Florencio-Martinez LE, Romero-Meza G, Rojas-Sanchez S, Moreno-Campos R, Arroyo R, et al. BRF1, a subunit of RNA polymerase III transcription factor TFIIIB, is essential for cell growth of Trypanosoma brucei. Parasitology. 2015;142:1563–73.
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–14.
Waugh B, Ghosh A, Bhattacharyya D, Ghoshal N, Banerjee R. In silico work flow for scaffold hopping in Leishmania. BMC Res Notes. 2014;7:802.
Wincker P, Ravel C, Blaineau C, Pagès M, Jauffret Y, Dedet JP, et al. The Leishmania genome comprises 36 chromosomes conserved across widely divergent human pathogenic species. Nucleic Acids Res. 1996;24:1688–94.
Wong AK. Molecular genetics of the parasitic protozoan Leishmania. Biochem Cell Biol. 1995;73:235–40.
Yaffe MB, Leparc GG, Lai J, Obata T, Volinia S, Cantley LC. A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat Biotechnol. 2001;19:348–53.
Zilberstein D. Proteomic analysis of posttranslational modifications using iTRAQ in Leishmania. Methods Mol Biol. 2015;1201:261–8.
Acknowledgements
The authors gratefully acknowledge the financial support of Department of Biotechnology (BCIL /NER-BPMC/2013) (DBT Twinning Program), New Delhi, India and University Grant Commission (UGC) (35/ 57/2009 (SR)), New Delhi, India to Madhumita Manna. For Network Project (Project NWP 0005) and J.C. Bose Fellowship (SB/S2/JCB-65/2014) to Syamal Roy and Council of Scientific and Industrial Research, New Delhi, India (ACK.NO. 113193/2k12/1) for CSIR SRFship to Supriya Khanra. The authors are also thankful to the DPI, Higher Education Dept. Govt. of West Bengal, the Principal, Barasat Govt. College, Kolkata, India and the Director, Indian Institute of Chemical Biology, Kolkata for their help and support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Khanra, S., Sarraf, N.R., Lahiry, S. et al. Leishmania genomics: a brief account. Nucleus 60, 227–235 (2017). https://doi.org/10.1007/s13237-017-0210-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13237-017-0210-y