Abstract
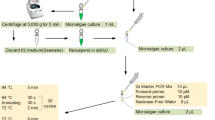
Countless species occur in the marine microalgal domain. Some are used as health functional foods or medical products but many species are harmful such as those that cause the red tide. Therefore, it is necessary to conduct prompt and accurate identification of microalgal species. As it is quite difficult to accurately distinguish all species in terms of morphology, we performed DNA barcoding analysis using molecular markers for more accurate and rapid screening. DNA barcoding analysis, i.e., DNA chip technology, is a powerful method for studies on microalgal taxonomy and biodiversity. We used the mitochondrial cytochrome c oxidase subunit I (mtCOI) as a barcoding gene to identify microalgal species. In this study, the diversity and phylogenetic differences among different microalgae were analyzed. Additionally, a microalgal species-specific probe was screened by 21–23 bp and the result was printed on silylated slide for use in a robotic microarrayer. As a result, we performed a DNA chip assay for each of 25 microalgal species and determined that the COI barcode gene was suitable as a marker gene, as it could identify various microalgae from the Korean South Sea by species.
Similar content being viewed by others
References
Kim, D. et al. Red to red-the marine bacterium Hahella chejuensis and its product prodigiosin for mitigation of harmful algal blooms. J. Microbiol. Biotechnol. 18, 1621–1629 (2008).
Hinder, S.L. et al. Toxic marine microalgae and shellfish poisoning in the British isles: history, review of epidemiology, and future implications. Environ. Health. 10, 54 (2011).
Moretti, V., Turchini, G., Bellagamba, F. & Caprino, F. Traceability issues in fishery and aquaculture products. Vet. Res. Commun. 27, 497–505 (2003).
Cunningham, E. & Meghen, C. Biological identification systems: genetic markers. Rev. Sci. Tech. 20, 491 (2001).
Ratnasingham, S., Hebert, P.D.N. & Gravenor, M.B., BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Mol. Ecol. Notes. 7, 355–364 (2007).
Yoon, H., Kim, G., Jeong, D., Jung, J. & Chung, I. Development of salmon identification DNA chip based on mitochondrial COIII-ND3-ND4L variations. Bio-Chip J. 2, 287–295 (2008).
Park, J.Y. et al. A DNA microarray for species identification of cetacean animals in Korean water. Bio-Chip J. 4, 197–203 (2010).
Kim, S. et al. DNA chip for species identification of Korean freshwater fish: A case study. BioChip J. 5, 72–77 (2011).
Hebert, P.D.N., Ratnasingham, S. & de Waard, J.R. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond., B, Biol. Sci. 270, S96 (2003).
Robba, L., Russell, S.J., Barker, G.L. & Brodie, J. Assessing the use of the mitochondrial cox1 marker for use in DNA barcoding of red algae (Rhodophyta). Am. J. Bot. 93, 1101–1108 (2006).
Alverson, A.J. & Kolnick, L. Intragenomic Nucleotide Polymorphism Among Small Subunit (18S) Rdna Paralogs in the Diatom Genus Skeletonema (Bacillariophyta) 1. J. Phycol. 41, 1248–1257 (2005).
Moniz, M.B.J., Kaczmarska, I. & Gravenor, M.B. Barcoding diatoms: Is there a good marker? Mol. Ecol. Resour. 9, 65–74 (2009).
Hebert, P.D. & Gregory, T.R. The promise of DNA barcoding for taxonomy. Syst. Biol. 54, 852–859 (2005).
Hajibabaei, M., Singer, G.A., Clare, E.L. & Hebert, P.D. Design and applicability of DNA arrays and DNA barcodes in biodiversity monitoring. BMC Biol. 5, 24 (2007).
Hebert, P.D.N., Cywinska, A., Ball, S.L. & DeWaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond., B, Biol. Sci. 270, 313 (2003).
Hajibabaei, M., Janzen, D.H., Burns, J.M., Hallwachs, W. & Hebert, P.D. DNA barcodes distinguish species of tropical Lepidoptera. Proc. Natl. Acad. Sci. U S A 103, 968–971 (2006).
Folmer, O., Black, M.A., Hoch, W., Lutz, R. & Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299 (1994).
Tamura, K., Dudley, J., Nei, M. & Kumar, S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596 (2007).
Engelmann, J.C. et al. Modelling cross hybridization on phylogenetic DNA microarrays increases the detection power of closely related species. Mol. Ecol. Resour. 9, 83–93 (2009).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lee, G., Park, S.Y., Yum, S. et al. Development of DNA chip for verification of 25 microalgae collected from southern coastal region in Korea. BioChip J 6, 325–334 (2012). https://doi.org/10.1007/s13206-012-6404-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13206-012-6404-0