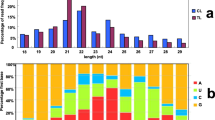
Abstract
MicroRNAs (miRNAs) play key regulatory roles in the plant’s response to biotic and abiotic stresses and have fundamental functions in plant–virus interactions. The study of changes in miRNAs in response to virus infection can provide molecular details for a better understanding of virus-host interactions. Maize Iranian mosaic virus (MIMV) infects maize and certain other poaceous plants but miRNA changes in response to MIMV infection are unknown. In the present study, we compared the miRNA profiles of MIMV-infected and uninfected maize and characterized their predicted roles in response to the virus. Small RNA sequencing of maize identified 257 conserved miRNAs of 26 conserved families in uninfected and MIMV-infected maize libraries. Among them, miR395, miR166 and miR156 family members were highly represented. Small RNA data were confirmed using RT-qPCR. In addition, 33 potential novel miRNAs were predicted. The data show that 13 miRNAs were up-regulated and 113 were down-regulated in response to MIMV infection. Several of those miRNAs are known to be important in the response to plant pathogens. To determine the potential roles of individual miRNAs in response to MIMV, miRNA targets, predicted interactions with circular RNAs and comparative transcriptome data were analyzed. The expression profiles of different miRNAs in response to MIMV provide novel insights into the roles of miRNAs in the interaction between MIMV and maize plants.




Similar content being viewed by others
References
Alvarado VY, Scholthof HB (2012) AGO2: a new Argonaute compromising plant virus accumulation. Front Plant Sci 2:112
Boyko A, Kathiria P, Zemp FJ, Yao Y, Pogribny I, Kovalchuk I (2007) Transgenerational changes in the genome stability and methylation in pathogen-infected plants: (virus-induced plant genome instability). Nucleic Acids Res 35:1714–1725
Cao M, Du P, Wang X, Yu Y-Q, Qiu Y-H, Li W, Gal-On A, Zhou C, Li Y, Ding S-W (2014) Virus infection triggers widespread silencing of host genes by a distinct class of endogenous siRNAs in Arabidopsis. Proc Natl Acad Sci 111:14613–14618
Chiniquy D, Underwood W, Corwin J, Ryan A, Szemenyei H, Lim CC, Stonebloom SH, Birdseye DS, Vogel J, Kliebenstein D (2019) PMR 5, an acetylation protein at the intersection of pectin biosynthesis and defense against fungal pathogens. Plant J 100:1022–1035
Cui J, You C, Chen X (2017) The evolution of microRNAs in plants. Curr Opin Plant Biol 35:61–67
Dai X, Zhuang Z, Zhao PX (2018) psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res 46:W49–W54
De Alba AEM, Elvira-Matelot E, Vaucheret H (2013) Gene silencing in plants: a diversity of pathways. Biochim Biophys Acta (BBA) Gene Regul Mech 1829:1300–1308
Díaz-Cruz GA, Cassone BJ (2018) A tale of survival: molecular defense mechanisms of soybean to overcome soybean mosaic virus infection. Physiol Mol Plant Pathol 102:79–87
Fahim M, Millar AA, Wood CC, Larkin PJ (2012) Resistance to wheat streak mosaic virus generated by expression of an artificial polycistronic microRNA in wheat. Plant Biotechnol J 10:150–163
Ghorbani A, Izadpanah K, Dietzgen RG (2018a) Changes in maize transcriptome in response to maize Iranian mosaic virus infection. PLoS ONE 13:e0194592
Ghorbani A, Izadpanah K, Dietzgen RG (2018b) Completed sequence and corrected annotation of the genome of maize Iranian mosaic virus. Adv Virol 163:767–770
Ghorbani A, Izadpanah K, Dietzgen RG (2018c) Gene expression and population polymorphism of maize Iranian mosaic virus in Zea mays, and intracellular localization and interactions of viral N, P, and M proteins in Nicotiana benthamiana. Virus Genes 54:290–296
Ghorbani A, Izadpanah K, Peters JR, Dietzgen RG, Mitter N (2018d) Detection and profiling of circular RNAs in uninfected and maize Iranian mosaic virus-infected maize. Plant Sci 274:402–409
Girardi E, López P, Pfeffer S (2018) On the importance of host microRNAs during viral infection. Front Genet 9:439
Iqbal MS, Jabbar B, Sharif MN, Ali Q, Husnain T, Nasir IA (2017) In silico MCMV silencing concludes potential host-derived miRNAs in maize. Front Plant Sci 8:372
Izadpanah K, Ahmadi A, Parvin S, Jafari S (1983) Transmission, particle size and additional hosts of the rhabdovirus causing maize mosaic in Shiraz, Iran 1. J Phytopathol 107:283–288
Jiang S, Lu Y, Li K, Lin L, Zheng H, Yan F, Chen J (2014) Heat shock protein 70 is necessary for rice stripe virus infection in plants. Mol Plant Pathol 15:907–917
Khalid A, Zhang Q, Yasir M, Li F (2017) Small RNA based genetic engineering for plant viral resistance: application in crop protection. Front Microbiol 8:43
Lei J, Sun Y (2014) miR-PREFeR: an accurate, fast and easy-to-use plant miRNA prediction tool using small RNA-Seq data. Bioinformatics 30:2837–2839
Leyser O (2018) Auxin signaling. Plant Physiol 176:465–479
Li Y, Lu Y-G, Shi Y, Wu L, Xu Y-J, Huang F, Guo X-Y, Zhang Y, Fan J, Zhao J-Q (2014) Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol 164:1077–1092
Li A, Li G, Zhao Y, Meng Z, Zhao M, Li C, Zhang Y, Li P, Ma C-L, Xia H (2018) Combined small RNA and gene expression analysis revealed roles of miRNAs in maize response to rice black-streaked dwarf virus infection. Sci Rep 8:1–14
Liu S-R, Zhou J-J, Hu C-G, Wei C-L, Zhang J-Z (2017) MicroRNA-mediated gene silencing in plant defense and viral counter-defense. Front Microbiol 8:1801
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Lu S, Li Q, Wei H, Chang M-J, Tunlaya-Anukit S, Kim H, Liu J, Song J, Sun Y-H, Yuan L (2013) Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc Natl Acad Sci 110:10848–10853
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338
Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312:436–439
Pantano L, Pantano F, Marti E, Sui SH (2019) Visualization of the small RNA transcriptome using seqclusterViz. F1000Research 8. https://doi.org/10.12688/f1000research.18142.1
Peace RJ, Hassani MS, Green JR (2019) mipIe: NGs-based prediction of miRNA using integrated evidence. Sci Rep 9:1548
Scheel TK, Luna JM, Liniger M, Nishiuchi E, Rozen-Gagnon K, Shlomai A, Auray G, Gerber M, Fak J, Keller I (2016) A broad RNA virus survey reveals both miRNA dependence and functional sequestration. Cell Host Microbe 19:409–423
Shivaprasad PV, Chen H-M, Patel K, Bond DM, Santos BA, Baulcombe DC (2012) A microRNA superfamily regulates nucleotide binding site–leucine-rich repeats and other mRNAs. Plant Cell 24:859–874
Tabara M, Ohtani M, Kanekatsu M, Moriyama H, Fukuhara T (2018) Size distribution of small interfering RNAs in various organs at different developmental stages is primarily determined by the dicing activity of dicer-like proteins in plants. Plant Cell Physiol 59:2228–2238
Trobaugh DW, Gardner CL, Sun C, Haddow AD, Wang E, Chapnik E, Mildner A, Weaver SC, Ryman KD, Klimstra WB (2014) RNA viruses can hijack vertebrate microRNAs to suppress innate immunity. Nature 506:245–248
Várallyay É, Válóczi A, Ágyi Á, Burgyán J, Havelda Z (2010) Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO J 29:3507–3519
Webb CA, Richter TE, Collins NC, Nicolas M, Trick HN, Pryor T, Hulbert SH (2002) Genetic and molecular characterization of the maize rp3 rust resistance locus. Genetics 162:381–394
Wu R, Wood M, Thrush A, Walton EF, Varkonyi-Gasic E (2007) Real-time PCR quantification of plant miRNAs using universal ProbeLibrary technology. Biochem-Mannh 2:12
Xia Z, Zhao Z, Li M, Chen L, Jiao Z, Wu Y, Zhou T, Yu W, Fan Z (2018) Identification of miRNAs and their targets in maize in response to sugarcane mosaic virus infection. Plant Physiol Biochem 125:143–152
Xia Z, Zhao Z, Gao X, Jiao Z, Wu Y, Zhou T, Fan Z (2019) Characterization of maize miRNAs in response to synergistic infection of maize chlorotic mottle virus and sugarcane mosaic virus. Int J Mol Sci 20:3146
Xu D, Mou G, Wang K, Zhou G (2014) MicroRNAs responding to southern rice black-streaked dwarf virus infection and their target genes associated with symptom development in rice. Virus Res 190:60–68
Zhou Y, Xu Z, Duan C, Chen Y, Meng Q, Wu J, Hao Z, Wang Z, Li M, Yong H (2016) Dual transcriptome analysis reveals insights into the response to rice black-streaked dwarf virus in maize. J Exp Bot 67:4593–4609
Acknowledgements
AG was supported by a fellowship from the National Foundation of Elites, Shiraz University and Iranian Council of Centers of Excellence. This research was jointly supported by the University of Queensland and the Queensland Department of Agriculture and Fisheries. Fund for Small RNA-Seq and preliminary experiments was provided by Plant Virology Research Center, College of Agriculture, Shiraz University, Iran.
Author information
Authors and Affiliations
Contributions
AG, KI, AA and RD designed the study; AG performed the experiments, AG, AT and AM analyzed the data and drafted the manuscript, all authors edited and approved the final version of the manuscripts.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Additional information
Accession numbers: SAMN15248610 and SAMN15248611 (NCBI).
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ghorbani, A., Izadpanah, K., Tahmasebi, A. et al. Characterization of maize miRNAs responsive to maize Iranian mosaic virus infection. 3 Biotech 12, 69 (2022). https://doi.org/10.1007/s13205-022-03134-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-022-03134-1