Abstract
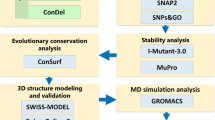
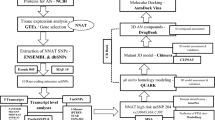
Neuronal growth regulator 1 (NEGR1) is a candidate gene for human obesity, which encodes the neural cell adhesion and growth molecule. The aim of the current study was to recognize the non-synonymous SNPs (nsSNPs) with the highest predicted deleterious effect on protein function of the NEGR1 gene. We have used five computational tools, namely, PolyPhen, SIFT, PROVEAN, MutPred and M-CAP, to predict the deleterious and pathogenic nsSNPs of the NEGR1 gene. Homology modeling approach was used to model the native and mutant NEGR1 protein models. Furthermore, structural validation was performed by the PROCHECK server to interpret the stability of the predicted models. We have predicted four potential deleterious nsSNPs, i.e., rs145524630 (Ala70Thr), rs267598710 (Pro168Leu), rs373419972 (Arg239Cys) and rs375352213 (Leu158Phe), which might be involved in causing obesity phenotypes. The predicted mutant models showed higher root mean square deviation and free energy values under the PyMoL and SWISS-PDB viewer, respectively. Additionally, the FTSite server predicted one nsSNP, i.e., rs145524630 (Ala70Thr) out of four identified nsSNPs found in the NEGR1 protein-binding site. There were four potential deleterious and pathogenic nsSNPs, i.e., rs145524630, rs267598710, rs373419972 and rs375352213, identified from the above-mentioned tools. In future, further functional in vitro and in vivo analysis could lead to better knowledge about these nsSNPs on the influence of the NEGR1 gene in causing human obesity. Hence, the present computational examination suggest that predicated nsSNPs may feasibly be a drug target and play an important role in contributing to human obesity.





Similar content being viewed by others
References
Adzhubei IA, Schmidt S, Peshkin L et al (2010) A method and server for predicting damaging missense mutations. Nat Methods 7:248–249. https://doi.org/10.1038/nmeth0410-248
Agalliu I, Williams S, Adler B et al (2015) The impact of obesity on prostate cancer recurrence observed after exclusion of diabetics. Cancer Causes Control 26:821–830. https://doi.org/10.1007/s10552-015-0554-z
Alonso R, Farías M, Alvarez V, Cuevas A (2016) The genetics of obesity. In: Translational cardiometabolic genomic medicine. Elsevier, Amsterdam, pp 161–177
Altschul SF, Madden TL, Schäffer AA et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201. https://doi.org/10.1093/bioinformatics/bti770
Biasini M, Bienert S, Waterhouse A et al (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. https://doi.org/10.1093/nar/gku340
Bruccoleri RE, Karplus M (1990) Conformational sampling using high-temperature molecular dynamics. Biopolymers 29:1847–1862. https://doi.org/10.1002/bip.360291415
Choi Y, Sims GE, Murphy S et al (2012) Predicting the functional effect of amino acid substitutions and indels. PLoS One 7:e46688. https://doi.org/10.1371/journal.pone.0046688
Dario AB, Ferreira ML, Refshauge KM et al (2015) The relationship between obesity, low back pain, and lumbar disc degeneration when genetics and the environment are considered: a systematic review of twin studies. Spine J 15:1106–1117. https://doi.org/10.1016/j.spinee.2015.02.001
Delorenzi M, Speed T (2002) An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics 18:617–625
Finucane MM, Stevens GA, Cowan MJ et al (2011) National, regional, and global trends in body-mass index since 1980: systematic analysis of health examination surveys and epidemiological studies with 960 country-years and 9·1 million participants. Lancet 377:557–567. https://doi.org/10.1016/S0140-6736(10)62037-5
Frayling TM, Timpson NJ, Weedon MN et al (2007) A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316:889–894. https://doi.org/10.1126/science.1141634
Gamero-Villarroel C, González LM, Gordillo I et al (2015) Impact of NEGR1 genetic variability on psychological traits of patients with eating disorders. Pharmacogenom J 15:278–283. https://doi.org/10.1038/tpj.2014.53
Garg ML, Wood LG (2013) Nutrition and physical activity in inflammatory diseases. CABI, Wallingford
Goswami AM (2015) Structural modeling and in silico analysis of non-synonymous single nucleotide polymorphisms of human 3β-hydroxysteroid dehydrogenase type 2. Meta Gene 5:162–172. https://doi.org/10.1016/j.mgene.2015.07.007
Griffith OL, Montgomery SB, Bernier B et al (2007) ORegAnno: an open-access community-driven resource for regulatory annotation. Nucleic Acids Res 36:D107–D113. https://doi.org/10.1093/nar/gkm967
Guh DP, Zhang W, Bansback N et al (2009) The incidence of co-morbidities related to obesity and overweight: a systematic review and meta-analysis. BMC Public Health. https://doi.org/10.1186/1471-2458-9-88
Hashimoto T, Yamada M, Maekawa S et al (2008) IgLON cell adhesion molecule Kilon is a crucial modulator for synapse number in hippocampal neurons. Brain Res 1224:1–11. https://doi.org/10.1016/j.brainres.2008.05.069
Jagadeesh K, Wenger A, Berger M, Guturu H, Stenson P, Cooper D, Bernstein J, Bejerano G (2016) M-CAP eliminates a majority of variants with uncertain significance in clinical exomes at high sensitivity. Nat Genet. https://doi.org/10.1038/ng.3703
Kim H, Chun Y, Che L et al (2017) The new obesity-associated protein, neuronal growth regulator 1 (NEGR1), is implicated in Niemann–Pick disease Type C (NPC2)-mediated cholesterol trafficking. Biochem Biophys Res Commun 482:1367–1374. https://doi.org/10.1016/j.bbrc.2016.12.043
Komar AA (2007) GENETICS: SNPs, silent but not invisible. Science 315:466–467. https://doi.org/10.1126/science.1138239
Kotsis V, Stabouli S, Papakatsika S et al (2010) Mechanisms of obesity-induced hypertension. Hypertens Res 33:386–393. https://doi.org/10.1038/hr.2010.9
Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol 305:567–580. https://doi.org/10.1006/jmbi.2000.4315 (Edited by F. Cohen)
Kumar P, Henikoff S, Ng PC (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4:1073–1081. https://doi.org/10.1038/nprot.2009.86
Kumar P, Singh RK, Mahalingam K (2015) In silico analysis of fat mass obesity associated (FTO) gene using computational algorithms. Int J Pharma Bio Sci 6:B589–B599
Laskowski RA, Rullmannn JA, MacArthur MW et al (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8:477–486
Lee AWS, Hengstler H, Schwald K et al (2012) Functional inactivation of the genome-wide association study obesity gene neuronal growth regulator 1 in mice causes a body mass phenotype. PLoS One 7:e41537. https://doi.org/10.1371/journal.pone.0041537
Lee H, Ash GI, Angelopoulos TJ et al (2015) Obesity-related genetic variants and their associations with physical activity. Sports Med Open 1:34. https://doi.org/10.1186/s40798-015-0036-6
Li B, Krishnan VG, Mort ME et al (2009) Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics 25:2744–2750. https://doi.org/10.1093/bioinformatics/btp528
Lindahl E, Azuara C, Koehl P, Delarue M (2006) NOMAD-Ref: visualization, deformation and refinement of macromolecular structures based on all-atom normal mode analysis. Nucleic Acids Res 34:W52–W56. https://doi.org/10.1093/nar/gkl082
Lodish H, Berk A, Zipursky SL et al (2000) Molecular cell biology, 4th edn. W. H. Freeman, San Francisco
Luchsinger JA, Cheng D, Tang MX et al (2012) Central obesity in the elderly is related to late-onset Alzheimer disease. Alzheimer Dis Assoc Disord 26:101–105. https://doi.org/10.1097/WAD.0b013e318222f0d4
Lyon HN, Hirschhorn JN (2005) Genetics of common forms of obesity: a brief overview. Am J Clin Nutr 82:215S–217S
Marg A, Sirim P, Spaltmann F et al (1999) Neurotractin, a novel neurite outgrowth-promoting Ig-like protein that interacts with CEPU-1 and LAMP. J Cell Biol 145:865–876
McCrindle BW (2015) Cardiovascular consequences of childhood obesity. Can J Cardiol 31:124–130. https://doi.org/10.1016/j.cjca.2014.08.017
Miyata S, Matsumoto N, Taguchi K et al (2003) Biochemical and ultrastructural analyses of IgLON cell adhesion molecules, Kilon and OBCAM in the rat brain. Neuroscience 117:645–658
NCD Risk Factor Collaboration (2016) Trends in adult body-mass index in 200 countries from 1975 to 2014: a pooled analysis of 1698 population-based measurement studies with 19·2 million participants. Lancet 387:1377–1396. https://doi.org/10.1016/S0140-6736(16)30054-X
Ng PC, Henikoff S (2003) SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res 31:3812–3814
Ngan C-H, Hall DR, Zerbe B et al (2012) FTSite: high accuracy detection of ligand binding sites on unbound protein structures. Bioinformatics 28:286–287. https://doi.org/10.1093/bioinformatics/btr651
Nogueiras R, Wiedmer P, Perez-Tilve D et al (2007) The central melanocortin system directly controls peripheral lipid metabolism. J Clin Investig 117:3475–3488. https://doi.org/10.1172/JCI31743
Nolan CJ, Damm P, Prentki M (2011) Type 2 diabetes across generations: from pathophysiology to prevention and management. Lancet 378:169–181. https://doi.org/10.1016/S0140-6736(11)60614-4
Pischedda F, Szczurkowska J, Cirnaru MD et al (2014) A cell surface biotinylation assay to reveal membrane-associated neuronal cues: Negr1 regulates dendritic arborization. Mol Cell Proteom 13:733–748. https://doi.org/10.1074/mcp.M113.031716
Power C, Pinto Pereira SM, Law C, Ki M (2014) Obesity and risk factors for cardiovascular disease and type 2 diabetes: investigating the role of physical activity and sedentary behaviour in mid-life in the 1958 British cohort. Atherosclerosis 233:363–369. https://doi.org/10.1016/j.atherosclerosis.2014.01.032
Punta M, Coggill PC, Eberhardt RY et al (2012) The Pfam protein families database. Nucleic Acids Res 40:D290–D301. https://doi.org/10.1093/nar/gkr1065
Ramachandran S, Kota P, Ding F, Dokholyan NV (2011) Automated minimization of steric clashes in protein structures. Proteins Struct Funct Bioinf 79:261–270. https://doi.org/10.1002/prot.22879
Rozman V, Kunej T (2018) Harnessing omics big data in nine vertebrate species by genome-wide prioritization of sequence variants with the highest predicted deleterious effect on protein function. OMICS 22:410–421. https://doi.org/10.1089/omi.2018.0046
Schäfer M, Bräuer AU, Savaskan NE et al (2005) Neurotractin/kilon promotes neurite outgrowth and is expressed on reactive astrocytes after entorhinal cortex lesion. Mol Cell Neurosci 29:580–590. https://doi.org/10.1016/j.mcn.2005.04.010
Shastry BS (2009) SNPs: impact on gene function and phenotype. In: Komar AA (ed) Single nucleotide polymorphisms. Humana Press, Totowa, pp 3–22
Sickmeier M, Hamilton JA, LeGall T et al (2007) DisProt: the database of disordered proteins. Nucleic Acids Res 35:D786–D793. https://doi.org/10.1093/nar/gkl893
Singh RK, Mahalingam K (2017) In silico approach to identify non-synonymous SNPs in human obesity related gene, MC3R (melanocortin-3-receptor). Comput Biol Chem 67:122–130. https://doi.org/10.1016/j.compbiolchem.2016.12.009
Singh RK, Kumar P, Mahalingam K (2016) In silico analysis of Snps in human obesity related gene, SLC6A14 (solute carrier family 6 (amino acid transporter), member 14). Int J Pharm Bio Sci 248–256
Singh RK, Kumar P, Mahalingam K (2017) Molecular genetics of human obesity: a comprehensive review. C R Biol 340:87–108. https://doi.org/10.1016/j.crvi.2016.11.007
Speliotes EK, Willer CJ, Berndt SI et al (2010) Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 42:937–948. https://doi.org/10.1038/ng.686
Thorleifsson G, Walters GB, Gudbjartsson DF et al (2009) Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet 41:18–24. https://doi.org/10.1038/ng.274
Vazquez-Martinez R, Gasman S (2014) The regulated secretory pathway in neuroendocrine cells. Front Endocrinol. https://doi.org/10.3389/fendo.2014.00048
Veerappa AM, Saldanha M, Padakannaya P, Ramachandra NB (2013) Family-based genome-wide copy number scan identifies five new genes of dyslexia involved in dendritic spinal plasticity. J Hum Genet 58:539–547. https://doi.org/10.1038/jhg.2013.47
Visser A, Ioan-Facsinay A, de Mutsert R et al (2014) Adiposity and hand osteoarthritis: the Netherlands Epidemiology of Obesity study. Arthr Res Ther 16:R19. https://doi.org/10.1186/ar4447
Willer CJ, Speliotes EK, Loos RJF et al (2009) Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet 41:25–34. https://doi.org/10.1038/ng.287
Williams LM (2012) Hypothalamic dysfunction in obesity. Proc Nutr Soc 71:521–533. https://doi.org/10.1017/S002966511200078X
Acknowledgements
All authors are grateful to the Vellore Institute of Technology, Vellore, Tamilnadu, India, for providing the facility to carry out the presented work. Mr. Permendra Kumar is thankful to the University Grants Commission (Government of India) for providing the Rajiv Gandhi National Fellowship-Senior Research Fellowship (UGC-RGNF-SRF). The authors acknowledge Mr. Rajan Kumar Singh (Teaching cum Research Assistant, SBST, Vellore Institute of Technology, Vellore) for his technical help and manuscript preparation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest for publishing this work.
Additional information
In memory of Prof. K. Mahalingam
Rights and permissions
About this article
Cite this article
Kumar, P., Mahalingam, K. In silico approach to identify non-synonymous SNPs with highest predicted deleterious effect on protein function in human obesity related gene, neuronal growth regulator 1 (NEGR1). 3 Biotech 8, 466 (2018). https://doi.org/10.1007/s13205-018-1463-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-018-1463-0