Abstract
The aim of this study was to identify the carbapenemases from clinical carbapenem-resistant Acinetobacter baumannii–calcoaceticus complex (CRABC) isolates and to assess their potential dissemination by conjugation and natural transformation. CRABC (n = 101) were collected consecutively from inpatients of the University Hospital of Monastir, Tunisia, from 2013 to 2016. Antimicrobial susceptibility was determined by the disk diffusion method and E-test. Carbapenemase-encoding genes were screened by PCR. Genotyping was performed by Pasteur MLST scheme. Isolates were resistant to all beta-lactams, fluoroquinolones and aminoglycosides while 80 and 90% were susceptible to tigecycline and colistin, respectively. Resistance and intermediate resistance to imipenem were 87 and 13%, respectively. The genes blaOXA-24-like, blaOXA-58-like, blaOXA-143-like, blaOXA-48-like, blaVIM, blaIMP, and blaKPC were not found. The blaOXA-51-like and blaOXA-23-like genes were present in 100 and 82.17% isolates, respectively. One isolate (< 1%) carried blaNDM-1 and blaOXA-51-like and belonged to Sequence Type 85 (ST85). Absence of transconjugants suggests a chromosomal location of NDM-1 determinant. The blaNDM-1 gene was inserted in a truncated form of Tn125, which may explain the absence of blaNDM-1 carrier-transformants. To our knowledge, this is the first report of the finding of NDM-positive A. baumannii in Tunisian territory. The study shows that despite the low prevalence and potential spread of NDM-1 enzyme among CRABC, continuous regional antimicrobial resistance surveillance and improved infection control measures are required in Tunisia to prevent further dissemination.
Similar content being viewed by others
Introduction
Acinetobacter baumannii–calcoaceticus complex are non-fermentative Gram-negative bacilli that includes A. baumannii, an opportunistic and emerging pathogen associated with nosocomial infections worldwide. It causes ventilator-associated pneumonia, bloodstream infections, urinary tract infections, wound infections and meningitis, especially in critically ill patients (Dijkshoorn et al. 2007).
Acinetobacter baumannii isolates are often resistant to many antimicrobial classes, including carbapenems. Carbapenems have been the mainstay of treatment for A. baumannii infections, but the widespread of carbapenem-resistant A. baumannii (CRAB) isolates has become an important health problem worldwide, leading to limited therapeutic options. Different mechanisms are responsible for carbapenem resistance in A. baumannii, namely reduced permeability of outer-membrane, efflux, and enzymatic hydrolysis. In A. baumannii, carbapenem resistance is mostly related to the production of carbapenem-hydrolyzing β-lactamases of Ambler class D (CHDLs) that include the oxacillinases families OXA-23, OXA-24/40, OXA-58, OXA-143, OXA-235, and the intrinsic oxacillinase blaOXA-51-like associated with ISAba1. It can also be mediated by Ambler class A β-lactamases (GES, KPC) and class B metallo-β lactamases (MBLs) (IMP, VIM and NDM) (Da Silva and Domingues 2016).
Among the MBLs, the NDM-type β-lactamases are emergent carbapenemases. NDM-1 determinant was initially identified in Klebsiella pneumoniae and Escherichia coli isolates recovered from a Swedish patient who was previously hospitalized in India (Yong et al. 2009). The blaNDM-1 rapidly disseminated in the world but India remains the major reservoir of NDM producers (Nordmann et al. 2011). Enterobacteriaceae are usually referred to as the main NDM-producers (van Duin 2017). Since the first description of NDM-1, 18 variants of the blaNDM have been identified (Bacterial Antimicrobial Resistance Reference Gene Database—https://www.ncbi.nlm.nih.gov/bioproject/PRJNA313047. Accessed on 6 July 2017). To date, only NDM-1 and NDM-2 were reported in A. baumannii (Da Silva and Domingues 2016).
In Tunisia, the dissemination of carbapenemases of Ambler class D in A. baumannii has been reported in a few studies (Mansour et al. 2008; Hammami et al. 2011, 2017; Charfi-Kessis et al. 2014; Chihi et al. 2016). In 2013, Bonnin et al. reported the finding of an A. baumannii isolate carrying the NDM-1 determinant isolated from a Tunisian patient hospitalized in France (Bonnin et al. 2013). Otherwise, and to our knowledge, the carbapenemase NDM-1 or other variants have never been reported in clinical A. baumannii in Tunisian territory. The main goals of the present study were to identify the carbapenemases in clinical isolates of the complex A. baumannii–calcoaceticus with reduced susceptibility to imipenem and to evaluate their potential dissemination by conjugation and natural transformation.
Materials and methods
Bacterial isolates and identification
A total of 101 non-duplicate A. baumannii–calcoaceticus complex non-susceptible to imipenem isolates were collected from clinical samples in the Laboratory of Microbiology at the UHMT, which is a tertiary care hospital with 858 beds. The clinical samples were obtained between 2013 and 2016 from patients hospitalized in different wards. Isolates were identified in the laboratory, using Api20E (BioMérieux), and identified as belonging to complex A. baumannii–A. calcoaceticus. PCR amplification of the blaOXA-51-like gene was used for presumptive identification of A. baumannii (Turton et al. 2006). The conserved rpoB gene of isolate Ab51 was amplified and sequenced (Stabvida) to further confirm the species (La Scola et al. 2006).
Susceptibility testing and screening for MBL-producing strains
Antibiotic susceptibility was determined on Mueller–Hinton agar using the standard disk diffusion procedure. The following antibiotics were tested: ticarcillin, ticarcillin/clavulanic acid, piperacillin, piperacillin/tazobactam, ceftazidime, cefpirome, imipenem, ciprofloxacin, gentamicin, tobramycin, amikacin, tigecycline and colistin. Results were interpreted according to the recommendations of the Comité de l’antibiogramme de la Société Française de Microbiologie (CA-SFM) (2013) (http://www.sfmmicrobiologie.org). For tigecycline, as there are no guidelines for the interpretation of the susceptibility to this antibiotic in A. baumannii, we followed the criteria previously described by Jones et al. (2007). The minimum inhibitory concentrations (MICs) of imipenem were determined using an E-test (AB BioMérieux) and the MIC values were interpreted according to the guidelines of CA-SFM guidelines (2013). All isolates were screened for metallo-β-lactamases production using the EDTA double disk synergy test and combined disc-test. Isolates were considered MBL positive if there was an increase of ≥ 7 mm in the zone of inhibition around the imipenem + EDTA disc as compared to imipenem disk alone (Yong et al. 2002).
Screening of carbapenemase genes by PCR
The carbapenemase-encoding genes of the Ambler classes A, B and D were screened by multiplex and single PCR, using specific primers for blaOXA-23-like, blaOXA-24-like, blaOXA-58-like, blaOXA-143-like, blaOXA-48-like, blaVIM, blaIMP, blaNDM and blaKPC as previously described (Yigit et al. 2001; Da Silva et al. 2002; Woodford et al. 2006; Poirel et al. 2011; Erturk et al. 2014; Cardoso et al. 2002). Presence of the ISAba1 insertion sequence upstream the blaOXA-23-like and blaOXA-51-like genes was also assessed by end-point PCR, combining one primer that binds in ISAba1 (Heritier et al. 2006) and one binding in the blaOXA-23-like (Woodford et al. 2006) and blaOXA-51-like (Turton et al. 2006) genes, respectively. All the primers used in this study are shown in Table 1.
Identification of the bla NDM variant, genetic environment and plasmid typing
The blaNDM gene is usually associated with the Tn125 transposon, where one copy of the ISAba125 is located upstream the carbapenemase gene (Poirel et al. 2012). We designed some primers and combined them with described primers to analyse the genetic environment. The primers IPIS_F and NDM-R (Table 1), binding in ISAba125 and blaNDM, respectively, were used for PCR amplification in the blaNDM-positive isolate. The amplicon was purified with Illustra ExoStart (GE Healthcare), following manufacturer’s instructions, and sequenced at Stabvida. Presence of the Tn125 was checked with primers IPIS_F and TRANSIS_R (Pfeifer et al. 2011), binding in the ISAba125, located on both ends of the transposon. Since a truncated isoform of Tn125 was also described in A. baumannii (Bonnin et al. 2012), the ΔTn125 was screened by PCR with primers IPIS_F and ISAba14_F and with NDM-F plus ISAba14_F (Table 1). All these PCRs were performed with the Phusion polymerase (ThermoFisher Scientific). Cycling conditions were: initial denaturation cycle at 98 °C for 30 s, followed by 30 cycles of denaturation at 98 °C for 10 s, annealing at 58 °C for 10 s, extension at 72 °C for 30 s per 1 kb of expected PCR product, and final extension at 72 °C for 5 min.
Plasmid typing was performed in the NDM-positive strain to identify the most common plasmid Inc groups, namely FIA, FIB, FIC, HI1, HI2, I1-Iγ, L/M, N, P, W, T, A/C, K, B/O, X, Y, F and FIIA (Carattoli et al. 2005).
Multilocus sequence typing
The multilocus sequence typing (MLST) technique was performed for the NDM-positive A. baumannii (Diancourt et al. 2010), using the internal fragments of seven housekeeping genes (cpn60, fusA, gltA, pyrG, recA, rplB and rpoB), according to the schemes available at Institute Pasteur’s MLST website (http://www.pasteur.fr/mlst). The PCR products were purified with Illustra ExoStart (GE Healthcare) sequenced at Stabvida. The sequences of the seven housekeeping genes were analyzed by comparing them to the MLST database (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Abaumannii.html).
Conjugation and natural transformation assays
Conjugation assays were performed in duplicate, with NDM-positive A. baumannii Ab51 as the donor cell and E. coli J53 (azide resistant) as the recipient by liquid mating broth method. Cultures of donor and recipient cells (1:10) were added to 5 ml of fresh Trypticase soy broth (TSB) and incubated overnight at 37 °C. Transconjugants were selected on Trypticase soy agar (TSA) plates containing sodium azide (150 µg/ml; Sigma) and cefotaxime (1.25 µg/ml; Sigma) to select for plasmid-encoded resistance. Acquisition of the blaNDM gene by natural transformation was tested with total free DNA (naked DNA) from A. baumannii Ab51 as donor and the clinical naturally competent strain A. baumannii A118 as the recipient (Ramirez et al. 2010), following the Wilharm and colleagues protocol (Wilharm et al. 2013). Selection of transformants was performed in Luria–Bertani (LB; Fluka) agar (Liofilchem) plates supplemented with cefotaxime 30 µg/ml or imipenem (Sigma) 4 µg/ml. Positive and negative controls were performed with homologous DNA and water, respectively. Two independent transformation assays were performed in triplicate.
Results
Origin and identification bacterial isolates
Of the 101 carbapenem-resistant Acinetobacter baumannii–calcoaceticus complex (CRABC) isolates, 51.48% were isolated from tracheal aspirate, 7.92% from bronchial aspirate and 3.96% from sputum. Other samples were from skin wounds (10.89%), 5.94% from catheters, 5.94% from blood, 3.96% from drains, 2.97% from urine, 2.97% from other non-specified sources, 0.99% from amniotic fluid, 0.99% from genital tract, 0.99% from liquid puncture (ascites, pleural effusion, pericardial effusion) and 0.99% from cerebrospinal fluid. Isolates were recovered from patients in various hospital units, including the intensive care unit (ICU) (70.29%), surgery (11.88%), internal medicine (9.9%), followed by gynecology and emergency (2.97% each), pediatric and from other departments (0.99% each).
All 101 isolates were identified at the hospital as belonging to the complex A. baumannii–A. calcoaceticus and carried the blaOXA-51-like, suggesting that they were A. baumannii (Turton et. 2006) since it is the most common multiresistant species found at hospital settings, which is assumed by the majority of the studies with numerous isolates. Isolates with specific characteristics, like carrying a metallo-β-lactamase gene, should be identified by sequencing conserved genes. For the blaNDM-positive strain, further confirmation was done by sequencing of the conserved rpoB gene since it has been shown that rarely some A. non-baumannii strains may carry blaOXA-51-like as well (Lee et al. 2009).
Antibiotic susceptibility testing
The results of antibiotic susceptibility testing revealed that the isolates were resistant to almost all antibiotics tested that included β–lactams, fluoroquinolones and aminoglycosides (Table 2). Ninety per cent and 80% were susceptible to colistin and tigecycline, respectively. The MICs of imipenem ranged from 4 to 32 mg/L; 87.13% of the isolates were resistant to imipenem and the remaining were classified as intermediate. These latter were considered as CRABC. In addition, the activity of β-lactamase was inhibited by EDTA solution in one strain (Ab51), suggesting the production of a MBL by this strain.
Identification of the carbapenem resistance genes
The genes blaOXA-24-like, blaOXA-58-like, blaOXA-143-like, blaOXA-48-like, blaVIM, blaIMP, and blaKPC were not detected. The blaOXA-51-like was present in all the isolates (100%) and 82.17% carried the gene blaOXA-23-like. The ISAba1 was located upstream the gene in 19.8% and 55.4% of the blaOXA-51-like and the blaOXA-23-like-carrying isolates, respectively. The gene blaNDM was detected in only one isolate, Ab51. This isolate did not carry any other tested acquired carbapenemase genes except blaOXA-51-like.
Characterization of the bla NDM-positive isolate
The sequencing results identified the blaNDM-1 variant in A. baumannii Ab51. This strain was isolated from a bronchial aspirate in a patient hospitalized in the ICU ward. The imipenem MIC was > 32 mg/L.
One plasmid was identified belonging to the replicon type IncW. Conjugation and natural transformation failed to produce transconjugants or transformants with blaNDM, respectively, as confirmed by PCR, both on cefotaxime or imipenem selection plates.
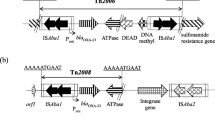
Figure 1 shows the genetic environment of the blaNDM gene and the strategy used for its detection. The blaNDM-1 gene was found embedded into ΔTn125. Genotyping by MLST revealed that the blaNDM-1 positive strain belongs to the sequence type 85 (ST85).
Discussion
Acinetobacter baumannii clinical isolates are usually multidrug resistant (MDR) and carbapenems have been used as one of the last therapeutic resort for the treatment of these MDR infections. The emergence of CRAB have been observed worldwide, including in countries of the Mediterranean basin (Mathlouthi et al. 2016). However, in Tunisia there is a limited knowledge in the epidemiology of clinical CRAB producing carbapenemases.
During the 4 years (2013–2016) of the study, 101 non-repetitive CRABC were collected from different patients at the UHMT. The majority of CRABC isolates were collected from inpatients of the ICU and from respiratory tract samples, but only a small percentage was isolated from blood samples. Our results differ from a previous study conducted in Tunisia (Hammami et al. 2011) that showed that CRAB strains were mostly isolated from blood cultures (32%), followed by pulmonary specimens (26%), with the majority of the samples collected from surgery ward (60%) and ICU (22%).
All the isolates were resistant or showed reduced susceptibility to imipenem. The frequency of resistance to amikacin, gentamicin, and ciprofloxacin was high and similar to previous findings in Tunisia (Hammami et al. 2011; Erturk et al. 2014). Amikacin is often reported as the aminoglycoside that shows better activity against MDR A. baumannii; yet, in Tunisia its efficacy may be compromised by the high-resistance rate. Tigecycline and colistin have been used for the treatment of infections caused by CRAB. However, we found already that 12 and 10% of the isolates were resistant to tigecycline and colistin, respectively. Previous studies in Tunisia, Libya and Turkey showed that all tested A. baumannii strains were susceptible to colistin (Mansour et al. 2008; Hammami et al. 2011; Erturk et al. 2014; Mathlouthi et al. 2016), which suggests the recent emergence of extended-drug resistant (XDR) strains.
In this study, OXA-23-like was found in the majority of CRABC, which is in accordance with the previous epidemiological studies performed in Tunisia (Mansour et al. 2008; Hammami et al. 2011, 2017; Charfi-Kessis et al. 2014). In Charles Nicolle Hospital of Tunisia, five A. baumannii isolates carried the OXA-23-like in association with GES-11 or GES-12 genes, but none of them harbored OXA-48-like or NDM determinants (Hammami et al. 2017). The ISAba1, associated with expression of the carbapenemase, was found located upstream the blaOXA-23-like in more than half the isolates. Other isolates carried the ISAba1 upstream blaOXA-51-like and/or may have other resistance mechanisms that contribute to carbapenem resistance. Here, we report for the first time the finding of the blaNDM-1 gene in A. baumannii in Tunisian territory. The frequency was rather low (1 out of 101 isolates during 4 years). In this country, the NDM-1 enzyme has only been reported in Enterobacteriaceae (Messaoudi et al. 2017). Nonetheless, NDM-1 has been found in other North African countries, such as Algeria (Boulanger et al. 2012), Libya (Mathlouthi et al. 2016) and Egypt (El-Sayed-Ahmed et al. 2015). This carbapenemase was also reported in six CRAB strains isolated in France from four patients with origin in Algeria, one in Tunisia and one in Egypt (Bonnin et al. 2013). However, in Tunisia the circulation of NDM-1-producing A. baumannii strains is unknown. Our work includes a considerable number of isolates (n = 101), and shows that, at least in this hospital, the isolation of NDM-1-producing A. baumannii strains is rare since only one isolate was identified. Despite the resistance of the strain, the infection could be controlled and the patient was discharged. No direct link could be established with CRAB NDM-producers of geographically close regions since the patient did not travel abroad. The NDM-1 producer Ab51 belongs to ST85, the same ST of the NDM-1 producer isolate recovered from a Tunisian patient in France in 2012 (Bonnin et al. 2013). Although ST2 is the most frequent ST in North Africa countries (Mathlouthi et al. 2016; El-Sayed-Ahmed et al. 2015; Bakour et al. 2015), a recent study in Libya showed a clonal diversity among CRAB. However, only two isolates belonged to ST85: one producing the NDM-1/OXA-23 (like the CRAB strain identified in France), and the other expressing only the NDM-1 metallo-β-lactamase (Mathlouthi et al. 2016), as the Ab51 of the present study.
The blaNDM have been found in diverse genera located in different replicon type plasmids, explaining their expanded dissemination (Da Silva and Domingues 2016). However, conjugation assays failed to originate transconjugants, suggesting a chromosomal location for blaNDM-1. Indeed, NDM-1/2 determinants have been found in the chromosome inserted in the composite transposon Tn125, supporting the hypothesis that Tn125 is the major vehicle of NDM dissemination (Poirel et al. 2012; Bontron et al. 2016). The movement of carbapenemase genes among A. baumannii populations is complex and it cannot be often explained by conjugation. Recently, transduction has been suggested as an intraspecies lateral mechanism for blaNDM-1 spread (Krahn et al. 2016). However, to our knowledge, natural transformation has never been tested for horizontal transfer of blaNDM. Only one study reports the transfer by natural transformation of a carbapenemase gene embedded in an integron (Domingues et al. 2012). This mechanism is dependent on the competence of the recipient cells. So far, only two studies report that clinical A. baumannii strains may express natural competence (Ramirez et al. 2010; Wilharm et al. 2013). Therefore, this mechanism cannot be ruled out and we hypothesized that blaNDM-1 could be transferred by natural transformation. Transformants were obtained in selection plates; however, none of the tested transformants harbored the blaNDM-1. The analysis of the genetic environment of blaNDM-1 in Ab51 confirmed its location in the truncated form of Tn125, as described previously in ten ST85 CRAB NDM-1-producers recovered in France between 2011 and 2012, being nine from North African patients (Bonnin et al. 2013, 2012). The absence of NDM-positive transformants might be explained by the presence of this truncated Tn125 in the Ab51, which prevents transposition to the genome of the recipient cells, as previously demonstrated for other functional transposons (Domingues et al. 2012). Based on previous results (Domingues et al. 2012), we may speculate that the non-truncated composite transposon Tn125 could transpose to competent A. baumannii recipient cells after uptake of DNA by natural transformation.
In conclusion, this study reports, to the best of our knowledge, the first occurrence of a A. baumannii NDM-1 producer in Tunisian territory, and reinforces the wide dissemination of OXA-23-like in this country. The low spread of blaNDM-1 among A. baumannii isolates might be related to a low mobilization due to its insertion in the truncated form of Tn125. This study allowed the implementation of more efficient control measures in this hospital. The travel of people across North Africa, South Europe, and East Mediterranean countries may contribute to the CRAB spread. In addition, the existence of ST85 A. baumannii strains carrying blaNDM-1 in North African neighbor countries, suggests possible clonal spread, probably by people cross-border dissemination. Therefore, it is imperative to develop antimicrobial stewardship initiatives and maintain antimicrobial resistance surveillance system in North Africa countries, as well as to screen patients in hospital admission to allow early detection and to prevent the high-dissemination of CRAB with application of convenient control measures.
References
Bakour S, Olaitan AO, Ammari H, Touati A, Saoudi S, Saoudi K, Rolain JM (2015) Emergence of colistin- and carbapenem-resistant Acinetobacter baumannii ST2 clinical isolate in Algeria: first case report. Microb Drug Resist 21(3):279–285. https://doi.org/10.1089/mdr.2014.0214
Bonnin RA, Poirel L, Naas T, Pirs M, Seme K, Schrenzel J, Nordmann P (2012) Dissemination of New Delhi metallo-beta-lactamase-1-producing Acinetobacter baumannii in Europe. Clin Microbiol Infect 18(9):E362–E365. https://doi.org/10.1111/j.1469-0691.2012.03928.x
Bonnin RA, Cuzon G, Poirel L, Nordmann P (2013) Multidrug-resistant Acinetobacter baumannii clone, France. Emerg Infect Dis 19(5):822–823. https://doi.org/10.3201/eid1905.121618
Bontron S, Nordmann P, Poirel L (2016) Transposition of Tn125 encoding the NDM-1 carbapenemase in Acinetobacter baumannii. Antimicrob Agents Chemother 60(12):7245–7251. https://doi.org/10.1128/AAC.01755-16
Boulanger A, Naas T, Fortineau N, Figueiredo S, Nordmann P (2012) NDM-1-producing Acinetobacter baumannii from Algeria. Antimicrob Agents Chemother 56(4):2214–2215. https://doi.org/10.1128/AAC.05653-11
Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ (2005) Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63(3):219–228. https://doi.org/10.1016/j.mimet.2005.03.018
Cardoso O, Leitao R, Figueiredo A, Sousa JC, Duarte A, Peixe LV (2002) Metallo-beta-lactamase VIM-2 in clinical isolates of Pseudomonas aeruginosa from Portugal. Microb Drug Resist 8(2):93–97. https://doi.org/10.1089/107662902760190635
Charfi-Kessis K, Mansour W, Ben Haj Khalifa A, Mastouri M, Nordmann P, Aouni M, Poirel L (2014) Multidrug-resistant Acinetobacter baumannii strains carrying the bla OXA-23 and the bla GES-11 genes in a neonatology center in Tunisia. Microb Pathog 74:20–24
Chihi H, Bonnin RA, Bourouis A, Mahrouki S, Besbes S, Moussa MB, Belhadj O, Naas T (2016) GES-11-producing Acinetobacter baumannii clinical isolates from Tunisian hospitals: Long-term dissemination of GES-type carbapenemases in North Africa. J Glob Antimicrob Resist 5:47–50. https://doi.org/10.1016/j.jgar.2016.03.005
Da Silva GJ, Domingues S (2016) Insights on the horizontal gene transfer of carbapenemase determinants in the opportunistic pathogen Acinetobacter baumannii. Microorganisms 4(3):23. https://doi.org/10.3390/microorganisms4030029
Da Silva GJ, Correia M, Vital C, Ribeiro G, Sousa JC, Leitão R, Peixe L, Duarte A (2002) Molecular characterization of bla IMP-5, a new integron-borne metallo-β-lactamase gene from Acinetobacter baumannii nosocomial isolate in Portugal. FEMS Microbiol Lett 215:33–39. https://doi.org/10.1111/j.1574-6968.2002.tb11366.x
Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S (2010) The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5(4):e10034. https://doi.org/10.1371/journal.pone.0010034
Dijkshoorn L, Nemec A, Seifert H (2007) An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol 5(12):939–951. https://doi.org/10.1038/nrmicro1789
Domingues S, Harms K, Fricke WF, Johnsen PJ, da Silva GJ, Nielsen KM (2012) Natural transformation facilitates transfer of transposons, integrons and gene cassettes between bacterial species. PLoS Pathog 8(8):e1002837. https://doi.org/10.1371/journal.ppat.1002837
El-Sayed-Ahmed MA, Amin MA, Tawakol WM, Loucif L, Bakour S, Rolain JM (2015) High prevalence of bla NDM–1 carbapenemase-encoding gene and 16S rRNA armA methyltransferase gene among Acinetobacter baumannii clinical Isolates in Egypt. Antimicrob Agents Chemother 59(6):3602–3605. https://doi.org/10.1128/AAC.04412-14
Erturk A, Cicek AC, Gumus A, Cure E, Sen A, Kurt A, Karagoz A, Aydogan N, Sandalli C, Durmaz R (2014) Molecular characterisation and control of Acinetobacter baumannii isolates resistant to multi-drugs emerging in inter-intensive care units. Ann Clin Microbiol Antimicrob 13:36. https://doi.org/10.1186/s12941-014-0036-2
Hammami S, Ghozzi R, Saidani M, Ben Redjeb S (2011) Carbapenem-resistant Acinetobacter baumannii producing the carbapenemase OXA-23 in Tunisia. Tunis Med 89(7):638–643
Hammami S, Dahdeh C, Mamlouk K, Ferjeni S, Maamar E, Hamzaoui Z, Saidani M, Ghedira S, Houissa M, Slim A, Boutiba-Ben Boubaker I (2017) Rectal carriage of extended-spectrum beta-lactamase and carbapenemase producing Gram-negative bacilli in intensive care units in Tunisia. Microb Drug Resist. https://doi.org/10.1089/mdr.2016.0205
Heritier C, Poirel L, Nordmann P (2006) Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii. Clin Microbiol Infect 12(2):123–130. https://doi.org/10.1111/j.1469-0691.2005.01320.x
Higgins PG, Poirel L, Lehmann M, Nordmann P, Seifert H (2009) OXA-143, a novel carbapenem-hydrolyzing class D beta-lactamase in Acinetobacter baumannii. Antimicrob Agents Chemother 53(12):5035–5038. https://doi.org/10.1128/AAC.00856-09
Jones RN, Ferraro MJ, Reller LB, Schreckenberger PC, Swenson JM, Sader HS (2007) Multicenter studies of tigecycline disk diffusion susceptibility results for Acinetobacter spp. J Clin Microbiol 45(1):227–230. https://doi.org/10.1128/JCM.01588-06
Krahn T, Wibberg D, Maus I, Winkler A, Bontron S, Sczyrba A, Nordmann P, Pühler A, Poirel L, Schlüter A (2016) Intraspecies transfer of the chromosomally encoded Acinetobacter baumannii bla NDM-1 carbapenemase gene. Antimicrob Agents Chemother. https://doi.org/10.1128/aac.00124-16
La Scola B, Gundi VA, Khamis A, Raoult D (2006) Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J Clin Microbiol 44(3):827–832. https://doi.org/10.1128/JCM.44.3.827-832.2006
Lee YT, Turton JF, Chen TL, Wu RC, Chang WC, Fung CP, Chen CP, Cho WL, Huang LY, Siu LK (2009) First identification of blaOXA-51-like in non-baumannii Acinetobacter spp. J Chemother 21(5):514–520. https://doi.org/10.1179/joc.2009.21.5.514
Mansour W, Poirel L, Bettaieb D, Bouallegue O, Boujaafar N, Nordmann P (2008) Dissemination of OXA-23-producing and carbapenem-resistant Acinetobacter baumannii in a University Hospital in Tunisia. Microb Drug Resist 14(4):289–292. https://doi.org/10.1089/mdr.2008.0838
Mathlouthi N, El Salabi AA, Ben Jomaa-Jemili M, Bakour S, Al-Bayssari C, Zorgani AA, Kraiema A, Elahmer O, Okdah L, Rolain JM, Chouchani C (2016) Early detection of metallo-beta-lactamase NDM-1- and OXA-23 carbapenemase-producing Acinetobacter baumannii in Libyan hospitals. Int J Antimicrob Agents 48(1):46–50. https://doi.org/10.1016/j.ijantimicag.2016.03.007
Messaoudi A, Haenni M, Mansour W, Saras E, Bel Haj Khalifa A, Chaouch C, Naija W, Boujaafar N, Bouallegue O, Madec JY (2017) ST147 NDM-1-producing Klebsiella pneumoniae spread in two Tunisian hospitals. J Antimicrob Chemother 72(1):315–316. https://doi.org/10.1093/jac/dkw401
Nordmann P, Poirel L, Walsh TR, Livermore DM (2011) The emerging NDM carbapenemases. Trends Microbiol 19(12):588–595. https://doi.org/10.1016/j.tim.2011.09.005
Pfeifer Y, Wilharm G, Zander E, Wichelhaus TA, Gottig S, Hunfeld KP, Seifert H, Witte W, Higgins PG (2011) Molecular characterization of bla NDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J Antimicrob Chemother 66(9):1998–2001. https://doi.org/10.1093/jac/dkr256
Poirel L, Heritier C, Tolun V, Nordmann P (2004) Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother 48(1):15–22
Poirel L, Walsh TR, Cuvillier V, Nordmann P (2011) Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70(1):119–123. https://doi.org/10.1016/j.diagmicrobio.2010.12.002
Poirel L, Bonnin RA, Boulanger A, Schrenzel J, Kaase M, Nordmann P (2012) Tn125-related acquisition of bla NDM-like genes in Acinetobacter baumannii. Antimicrob Agents Chemother 56(2):1087–1089. https://doi.org/10.1128/AAC.05620-11
Ramirez MS, Don M, Merkier AK, Bistue AJ, Zorreguieta A, Centron D, Tolmasky ME (2010) Naturally competent Acinetobacter baumannii clinical isolate as a convenient model for genetic studies. J Clin Microbiol 48(4):1488–1490. https://doi.org/10.1128/JCM.01264-09
Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL (2006) Identification of Acinetobacter baumannii by detection of the bla OXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol 44(8):2974–2976. https://doi.org/10.1128/JCM.01021-06
van Duin D (2017) Carbapenem-resistant Enterobacteriaceae: what we know and what we need to know. Virulence 8(4):379–382. https://doi.org/10.1080/21505594.2017.1306621
Wilharm G, Piesker J, Laue M, Skiebe E (2013) DNA uptake by the nosocomial pathogen Acinetobacter baumannii occurs during movement along wet surfaces. J Bacteriol 195(18):4146–4153. https://doi.org/10.1128/JB.00754-13
Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, Amyes SG, Livermore DM (2006) Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents 27(4):351–353. https://doi.org/10.1016/j.ijantimicag.2006.01.004
Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC (2001) Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45(4):1151–1161. https://doi.org/10.1128/AAC.45.4.1151-1161.2001
Yong D, Lee K, Yum JH, Shin HB, Rossolini GM, Chong Y (2002) Imipenem-EDTA disk method for differentiation of metallo-beta-lactamase-producing clinical isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol 40(10):3798–3801. https://doi.org/10.1128/JCM.40.10.3798-3801.2002
Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR (2009) Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53(12):5046–5054. https://doi.org/10.1128/AAC.00774-09
Acknowledgements
H. Ben Cheikh was supported by the grant of the Ministry of Higher Education and Research of Tunisia.
Funding
The study was supported by funds of the Faculty of Pharmacy of the University of Coimbra, Coimbra, Portugal; “Fundos FEDER, Programa Operacional Factores de Competitividade-COMPETE 2020 and Fundos Nacionais através da FCT-Fundação para a Ciência e a Tecnologia, projecto Estratégico COMPETE: POCI-01-0145-FEDER-007440”, through the Center for Neuroscience and Cell Biology, University of Coimbra, Portugal; and the Laboratory of Biologically Active Substances and Communicable Diseases (LR99ES27), Monastir University, the Fattouma Bourguiba University Hospital, Monastir, Tunisia and Faculty of Science, Bizerta Carthage University, Bizerta, Tunisia.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest.
Rights and permissions
About this article
Cite this article
Cheikh, H.B., Domingues, S., Silveira, E. et al. Molecular characterization of carbapenemases of clinical Acinetobacter baumannii–calcoaceticus complex isolates from a University Hospital in Tunisia. 3 Biotech 8, 297 (2018). https://doi.org/10.1007/s13205-018-1310-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-018-1310-3