Abstract
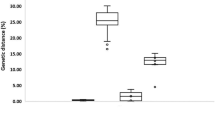
Identification of hydrozoan species is challenging, even for taxonomic experts, due to the scarcity of distinct morphological characters and phenotypic plasticity. DNA barcoding provides an efficient method for species identification, however, the choice between mitochondrial cytochrome c oxidase subunit I (COI) and large subunit ribosomal RNA gene (16S) as a standard barcode for hydrozoans is subject to debate. Herein, we directly compared the barcode potential of COI and 16S in hydrozoans using 339 sequences from 47 pelagic hydrozoan species. Analysis of Kimura 2-parameter genetic distances (K2P) documented the mean intraspecific/interspecific variation for COI and 16S to be 0.004/0.204 and 0.003/0.223, respectively. An obvious “barcoding gap” was detected for all species in both markers and all individuals of a species clustered together in both the COI and 16S trees. These results suggested that the species within the studied taxa can be efficiently and accurately identified by COI and 16S. Furthermore, our results confirmed that 16S was a better phylogenetic marker for hydrozoans at the genus level, and in some cases at the family level. Considering the resolution and effectiveness for barcoding and phylogenetic analyses of Hydrozoa, we strongly recommend 16S as the standard barcode for hydrozoans.
Similar content being viewed by others
References
Bouillon J, Boero F. 2000. The Hydrozoa: A new classification in the light of old knowledge. Thalassia Salentina, 24: 1–45
Bridge D, Cunningham C W, DeSalle R, et al. 1995. Class-level relationships in the Phylum Cnidaria: molecular and morphological evidence. Molecular Biology and Evolution, 12: 679–689
Bucklin A, Ortman B D, Jennings R M, et al. 2010. A “Rosetta Stone” for zooplankton: DNA barcode analysis of holozooplankton diversity of the Sargasso Sea (NW Atlantic Ocean). Deep-Sea Research II, 57: 2234–2247
Bucklin A, Steinke D, Blanco-Bercial L. 2011. DNA barcoding of marine metazoa. Annual Review of Marine Science, 3: 471–508
Cantero A L P, Sentandreu V, Latorre A. 2010. Phylogenetic relationships of the endemic Antarctic benthic hydroids (Cnidaria, Hydrozoa): what does the mitochondrial 16S rRNA tell us about it? Polar Biology, 33: 41–57
Cartwright P, Evans N M, Dunn C W, et al. 2008. Phylogenetics of Hydroidolina (Hydrozoa: Cnidaria). Journal of the Marine Biological Association of the United Kingdom, 88: 1663–1672
Collins A G. 2000. Towards understanding the phylogenetic history of Hydrozoa: hypothesis testing with 18S gene sequence data. Scientia Marina, 64: 5–22
Collins A G. 2002. Phylogeny of Medusozoa and the evolution of cnidarian life cycles. Journal of Evolutionary Biology, 15: 418–432
Collins A G, Bentlage B, Lindner A, et al. 2008. Phylogenetics of Trachylina (Cnidaria: Hydrozoa) with new insights on the evolution of some problematic taxa. Journal of the Marine Biological Association of the United Kingdom, 88: 1673–1685
Collins A G, Schuchert P, Marques A C, et al. 2006. Medusozoan phylogeny and character evolution clarified by new large and small subunit rDNA data and an assessment of the utility of phylogenetic mixture models. Systematic Biology, 55: 97–115
Collins A G, Winkelmann S, Hadrys H, et al. 2005. Phylogeny of Capitata and Corynidae (Cnidaria, Hydrozoa) in light of mitochondrial 16S rDNA data. Zoologica Scripta, 34: 91–99
del-Prado R, Cubas P, Lumbsch H T, et al. 2010. Genetic distances within and among species in monophyletic lineages of Parmeliceae (Ascomycota) as a tool for taxon delimitation. Molecular Phylogenetics and Evolution, 56: 125–133
Dawson M N. 2003. Macro-morphological variation mong cryptic specie of the moon jellyfish, Aurelia (Cnidaria: Scyphozoa). Marine Biology, 143: 369–379
Dawson M N. 2005. Cyanea capillata is not a cosmopolitan jellyfish: morphological and molecular evidence for C. annaskala and C. rosea (Scyphozoa: Semaeostomeae: Cyaneidae) in south-eastern Australia. Invertebrate Systematics, 19: 361–370
Dawson M N, Jacobs D K. 2001. Molecular evidence for cryptic species of Aurelia aurita (Cnidaria, Scyphozoa). Biological Bulletin, 200: 92–96
Dawson M N, Martin L E. 2001. Geographic variation and ecological adaptation in Aurelia (Scyphozoa, Semaeostomeae): some implications from molecular phylogenetics. Hydrobiologia, 451: 259–273
Dunn C W, Pugh P R, Haddock S H D. 2005. Molecular phylogenetics of the siphonophora (Cnidaria), with implications for the evolution of functional specialization. Systematic Biology, 54: 916–935
Ender A, Schierwater B. 2003. Placozoa are not derived cnidarians: evidence from molecular morphology. Molecuar Biology Evolution, 20: 130–134
Faith D P, Williams K J. 2005. How large-scale DNA barcoding programs can boost biodiversity conservation planning: linking phylogenetic diversity (PD) analyses to the barcode of life database (BoLD). Abstract. In: Australian entomological society’s 36th AGM and scientific conference/7th invertebrate biodiversity and conservation conference/Australian systematics society, Canberra, Australia, 4–9 December 2005
Folino-Rorem N C, Darling J A, D’Ausilio C A. 2008. Genetic analysis reveals multiple cryptic invasive species of the hydrozoan genus Cordylophora. Biological Invasions, 11: 1869–1882
Folmer O, Black M, Hoeh W, et al. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology, 3: 294–299
Govindarajan A F, Boero F, Halanych K M. 2006. Phylogenetic analysis with multiple markers indicates repeated loss of the adult medusa stage in Campanulariidae (Hydrozoa, Cnidaria). Molecular Phylogenetics and Evolution, 38: 820–834
Govindarajan A F, Halanych K M, Cunningham C W. 2005a. Mitochondrial evolution and phylogeography in the hydrozoan Obelia geniculata (Cnidaria). Marine Biology, 146: 213–222
Govindarajan A F, Piraino S, Gravili C, et al. 2005b. Species identification of bivalve-inhabiting marine hydrozoans of the genus Eugymnanthea. Invertebrate Biology, 124: 1–10
Hajibabaei M, Janzen D H, Burns J M, et al. 2006. DNA barcodes distinguish species of tropical Lepidoptera. Proceedings of the National Academy of Sciences of the United States of America, 103: 968–971
Hajibabaei M, Singer G A C, Hebert P D N, et al. 2007. DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends in Genetics, 23: 167–172
Hebert P D N, Gregory T R. 2005. The promise of DNA barcoding for taxonomy. Systematic Biology, 54: 852–859
Hebert P D N, Penton E H, Burns J M, et al. 2004a. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proceedings of the National Academy of Science of the United States of America, 101: 14812–14817
Hebert P D N, Ratnasingham S, deWaard J R. 2003. Barcoding animal life: cytochrome c oxidase subunit I divergences among closely related species. Proceedings of the Royal Society B-Biological Sciences, 270: S96–S99
Hebert P D N, Stoeckle M Y, Zemlak T S, et al. 2004b. Identification of birds through DNA barcodes. PloS Biology, 2: 1657–1663
Hellberg M E. 2006. No variation and low synonymous substitution rates in coral mtDNA despite high nuclear variation. BMC Evolutionary Biology, 6: 1–8
Holland B S, Dawson M N, Crow G L, et al. 2004. Global phylogeography of Cassiopea (Scyphozoa: Rhizostomeae): molecular evidence for cryptic species and multiple invasions of the Hawaiian Islands. Marine Biology, 145: 1119–1128
Huang D, Meier R, Todd P A, et al. 2008. Slow mitochondrial COI sequence evolution at the base of the metazoan tree and its implications for DNA barcoding. Journal of Molecular Evolution, 66: 167–174
Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16: 111–120
Köhler F. 2007. From DNA taxonomy to barcoding: how a vague idea evolved into a biosystematic tool. Mitteilungen aus dem Museum für Naturkunde Berlin Zoologische Reihe, 83: 44–51
Krishnamurthy P K, Francis R A. 2012. A critical review on the utility of DNA barcoding in biodiversity conservation. Biodiversity and Conservation, 21: 1901–1919
Kubota S. 1983. Studies on life history and systematics of the Japanese commensal hydroids living in bivalves, with some reference to their evolution. Journal of the Faculty of Science Hokkaido University Series VI Zoology, 23: 296–402
Kubota S. 2000. Parallel, paedomorphic evolutionary processes of the bivalve-inhabiting hydrozoans (Leptomedusae, Eirenidae) deduced from morphology, life cycle and biogeography, with special reference to taxonomic treatment of Eugymnanthea. Scientia Marina, 64: 241–247
Larkin M A, Blackshields G, Brown N P, et al. 2007. Clustal W and clustal × version 2.0. Bioinformatics, 23: 2947–2948
Leclère L, Schuchert P, Cruaud C, et al. 2009. Molecular phylogenetics of Thecata (Hydrozoa, Cnidaria) reveals long-term maintenance of life history traits despite high frequency of recent character changes. Systematic Biology, 58: 509–526
Leclère L, Schuchert P, Manuel M. 2007. Phylogeny of the Plumularioidea (Hydrozoa, Leptothecata): evolution of colonial organization and life cycle. Zoologica Scripta, 36: 371–394
Le Goff-Vitry M C, Rogers A D, Baglow D. 2004. A deep sea slant on the molecular phylogeny of the Scleractinia. Molecular Phylogenetics and Evolution, 30: 167–177
Martinez D E, Iniguez A R, Percell K M, et al. 2010. Phylogeny and biogeography of Hydra (Cnidaria: Hydridae) using mitochondrial and nuclear DNA sequences. Molecular Phylogenetics and Evolution, 57: 403–410
McFadden C S, Benayahu Y, Pante E, et al. 2011. Limitations of mitochondrial gene barcoding in Octocorallia. Molecular Ecology Resources, 11: 19–31
McFadden C S, Tullis I, Hutchinson M B, et al. 2000. Rates of evolution of cnidarian mitochondrial genes. American Zoologist, 40: 1124–1124
Meyer C P, Paulay G. 2005. DNA barcoding: error rates based on comprehensive sampling. PLoS Biolog, 3: 2229–2238
Miglietta M P, Piraino S, Kubota S, et al. 2007. Species in the genus Turritopsis (Cnidaria, Hydrozoa): a molecular evaluation. Journal of Zoological Systematics and Evolutionary Research, 45: 11–19
Miglietta M P, Schuchert P, Cunningham C W. 2009. Reconciling genealogical and morphological species in a worldwide study of the Family Hydractiniidae (Cnidaria, Hydrozoa). Zoologica Scripta, 38: 403–430
Mills C E. 1995. Medusae, siphonophores, and ctenophores as planktivorous predators in changing global ecosystems. ICES Journal of Marine Science, 52: 575–581
Miranda L S, Collins A G, Marques A C. 2010. Molecules clarify a Cnidarian life cycle — the “Hydrozoan” Microhydrula limopsicola is an early life stage of the Staurozoan Haliclystus antarcticus. PLoS One, 5: e10182
Moritz C, Cicero C. 2004. DNA barcoding: promise and pitfalls. Plos Biology, 2: 1529–1531
Moura C J, Cunha M R, Porteiro F M, et al. 2011a. Polyphyly and cryptic diversity in the hydrozoan families Lafoeidae and Hebellidae (Cnidaria:Hydrozoa). Invertebrate Systematics, 25: 454–470
Moura C J, Cunha M R, Porteiro F M, et al. 2011b. The use of the DNA barcode gene 16S mRNA for the clarification of taxonomic problems within the family Sertulariidae (Cnidaria, Hydrozoa). Zoologica Scripta, 40: 520–537
Moura C J, Cunha M R, Porteiro F M, et al. 2012. A molecular phylogenetic appraisal of the systematics of the Aglaopheniidae (Cnidaria: Hydrozoa, Leptothecata) from the north-east Atlantic and west Mediterranean. Zoological Journal of the Linnean Society, 164: 717–727
Moura C J, Harris D J, Cunha M R, et al. 2008. DNA barcoding reveals cryptic diversity in marine hydroids (Cnidaria, Hydrozoa) from coastal and deep-sea environments. Zoologica Scripta, 37: 93–108
Nawrocki A M, Schuchert P, Cartwright P. 2010. Phylogenetics and evolution of Capitata (Cnidaria: Hydrozoa), and the status of the family Corynidae. Zoological Scripta, 39: 290–304
Ortman B D, Bucklin A, Pagès F, et al. 2010. DNA barcoding the Medusozoa using mtCOI. Deep-Sea Research II: Topical Studies in Oceanography, 57: 2148–2156
Pontin D R, Cruickshank R H. 2012. Molecular phylogenetics of the genus Physalia (Cnidaria: Siphonophora) in New Zealand coastal waters reveals cryptic diversity. Hydrobiologia, 686: 91–105
Ratnasingham S, Hebert P D N. 2007. BOLD: the barcode of life data system (http://www.barcodinglife.org). Molecular Ecology Notes, 7: 355–364
Schuchert P. 2005a. Species boundaries in the hydrozoan genus Coryne. Molecular Phylogenetics and Evolution, 36: 194–199
Schuchert P. 2005b. Rediscovery of Coryne fucicola (de Filippi, 1866) (Cnidaria: Hydrozoa). Cahiers de Biologie Marine, 46: 305–310
Schuchert P. 2006. The European athecate hydroids and their medusae (Hydrozoa, Cnidaria): Capitata Part 1. Revue Suisse de Zoologie, 113: 325–410
Schuchert P. 2007. The European athecate hydroids and their medusae (Hydrozoa, Cnidaria): Filifera Part 2. Revue Suisse de Zoologie, 114: 195–396
Schuchert P. 2008a. The European athecate hydroids and their medusae (Hydrozoa, Cnidaria): Filifera Part 3. Revue Suisse de Zoologie, 115: 221–302
Schuchert P. 2008b. The European athecate hydroids and their medusae (Hydrozoa, Cnidaria): Filifera Part 4. Revue Suisse de Zoologie, 115: 677–757
Schuchert P. 2009. The European athecate hydroids and their medusae (Hydrozoa, Cnidaria): Filifera Part 5. Revue Suisse de Zoologie, 116: 441–507
Schuchert P, Reiswig H M. 2006. Brinckmannia hexactinellidophila, n.genera, n. sp. a hydroid living in tissues of glass sponges of the reefs, fjords, and seamounts of Pacific Canada and Alaska. Canadian Journal of Zoology, 84: 564–572
Shearer T L, van Oppen M J H, Romano S L, et al. 2002. Slow mitochondrial DNA sequence evolution in the Anthozoa (Cnidaria). Molecular Ecology, 11: 2475–2487
Sinniger F, Reimer J D, Pawlowski J. 2008. Potential of DNA sequences to identify zoanthids (Cnidaria: Zoantharia). Zoological Science, 25: 1253–1260
Stampar S N, Maronna M M, Vermeij M J A, et al. 2012. Evolutionary diversification of banded tube-dwelling anemones (Cnidaria; Ceriantharia; Isarachnanthus) in the Atlantic Ocean. PLoS One, 7: e41091
Sun Yan, Li Qi, Kong Lingfeng, et al. 2012. DNA barcoding of Caenogastropoda along coast of China based on the COI gene. Molecular Ecology Resources, 12: 209–218
Tamura K, Peterson D, Peterson N, et al. 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28: 2731–2739
Ward R D, Zemlak T S, Innes B H, et al. 2005. DNA barcoding Australia’s fish species. Philosophical Transactions of the Royal Society B—Biological Sciences, 360: 1847–1857
Xia Yun, Gu Haifeng, Peng Rui, et al. 2012. COI is better than 16S rRNA for DNA barcoding Asiatic salamanders (Amphibia: Caudata: Hynobiidae). Molecular Ecology Resources, 12: 48–56
Zemlak T S, Ward R D, Connell A D, et al. 2009. DNA barcoding reveals overlooked marine fishes. Molecular Ecology Resources, 9: 237–242
Zheng Lianming, Lin Yuanshao, Li Shaojing, et al. 2009. Aequorea taiwanensis n. sp (Hydrozoa, Leptomedusae) and mtCOI sequence analysis for the genus Aequorea. Acta Oceanologica Sinica, 28: 109–115
Zhou Konglin, Zheng Lianming, He Jinru, et al. 2013. Detection of a new Clytia species (Cnidaria: Hydrozoa: Campanulariidae) with DNA barcoding and life cycle analyses. Journal of the Marine Biological Association of the United Kingdom, 93: 2075–2088
Author information
Authors and Affiliations
Corresponding author
Additional information
Foundation item: The National Natural Science Foundation of China under contract No. 41006078; the Fundamental Research Funds for the Central Universities under contract No. 2010121037; the Public Science and Technology Research Funds Projects of Ocean under contract Nos 201005012-3 and 201005015-5; the Natural Science Foundation of Fujian Province of China under contract No. 2011J05116.
Rights and permissions
About this article
Cite this article
Zheng, L., He, J., Lin, Y. et al. 16S rRNA is a better choice than COI for DNA barcoding hydrozoans in the coastal waters of China. Acta Oceanol. Sin. 33, 55–76 (2014). https://doi.org/10.1007/s13131-014-0415-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13131-014-0415-8