Abstract
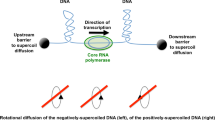
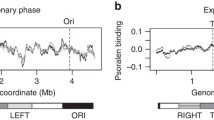
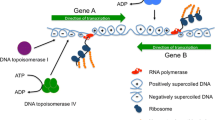
Bacteria organize DNA into self-adherent conglomerates called nucleoids that are replicated, transcribed, and partitioned within the cytoplasm during growth and cell division. Three classes of proteins help condense nucleoids: (1) DNA gyrase generates diffusible negative supercoils that help compact DNA into a dynamic interwound and multiply branched structure; (2) RNA polymerase and abundant small basic nucleoid-associated proteins (NAPs) create constrained supercoils by binding, bending, and forming cooperative protein–DNA complexes; (3) a multi-protein DNA condensin organizes chromosome structure to assist sister chromosome segregation after replication. Most bacteria have four topoisomerases that participate in DNA dynamics during replication and transcription. Gyrase and topoisomerase I (Topo I) are intimately involved in transcription; Topo III and Topo IV play critical roles in decatenating and unknotting DNA during and immediately after replication. RNA polymerase generates positive (+) supercoils downstream and negative (−) supercoils upstream of highly transcribed operons. Supercoil levels vary under fast versus slow growth conditions, but what surprises many investigators is that it also varies significantly between different bacterial species. The MukFEB condensin is dispensable in the high supercoil density (σ) organism Escherichia coli but is essential in Salmonella spp. which has 15 % fewer supercoils. These observations raise two questions: (1) How do different species regulate supercoil density? (2) Why do closely related species evolve different optimal supercoil levels? Control of supercoil density in E. coli and Salmonella is largely determined by differences encoded within the gyrase subunits. Supercoil differences may arise to minimalize toxicity of mobile DNA elements in the genome.




Similar content being viewed by others
References
Bacciu D, Falchi G, Spazziani A, Bossi L, Marogna G, Leori GS, Rubino S, Uzzau S (2004) Transposition of the heat-stable toxin astA gene into a gifsy-2-related prophage of Salmonella enterica serovar Abortusovis. J Bacteriol 186:4568–4574
Basu A, Schoeffler AJ, Berger JM, Bryant Z (2012) ATP binding controls distinct structural transitions of Escherichia coli DNA gyrase in complex with DNA. Nat Struct Mol Biol 19(538–546):S531
Booker BM, Deng S, Higgins NP (2010) DNA topology of highly transcribed operons in Salmonella enterica serovar Typhimurium. Mol Microbiol 78:1348–1364
Brandis G, Hughes D (2016) The selective advantage of synonymous codon usage bias in Salmonella. PLoS Genet 12:e1005926
Cagliero C, Zhou YN, Jin DJ (2014) Spatial organization of transcription machinery and its segregation from the replisome in fast-growing bacterial cells. Nucleic Acids Res 42:13696–13705
Champion K, Higgins NP (2007) Growth rate toxicity phenotypes and homeostatic supercoil control differentiate Escherichia coli from Salmonella enterica serovar Typhimurium. J Bacteriol 189:5839–5849
Chong S, Chen C, Ge H, Xie XS (2014) Mechanism of transcriptional bursting in bacteria. Cell 158:314–326
Coombes BK, Wickham ME, Brown NF, Lemire S, Bossi L, Hsiao WW, Brinkman FS, Finlay BB (2005) Genetic and molecular analysis of GogB, a phage-encoded type III-secreted substrate in Salmonella enterica serovar typhimurium with autonomous expression from its associated phage. J Mol Biol 348:817–830
Cunha S, Woldringh CL, Odijk T (2005) Restricted diffusion of DNA segments within the isolated Escherichia coli nucleoid. J Struct Biol 150:226–232
Datta S, Costantino N, Court DL (2006) A set of recombineering plasmids for gram-negative bacteria. Gene 379:109–115
Dillon SC, Dorman CJ (2010) Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol 8:185–195
Drolet M, Broccoli S, Rallu F, Hraiky C, Fortin C, Masse E, Baaklini I (2003) The problem of hypernegative supercoiling and R-loop formation in transcription. Front Biosci 8:D210–D221
Espeli O, Boccard F (1997) In vivo cleavage of Escherichia coli BIME-2 repeats by DNA gyrase: genetic characterization of the target and identification of the cut site. Mol Microbiol 26:767–777
Espeli O, Mercier R, Boccard F (2008) DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Mol Microbiol 68:1418–1427
Fernandez-Sierra M, Shao Q, Fountain C, Finzi L, Dunlap D (2015) E. coli gyrase fails to negatively supercoil diaminopurine-substituted DNA. J Mol Biol 427:2305–2318
Figueroa-Bossi N, Bossi L (1999) Inducible prophages contribute to Salmonella virulence in mice. Mol Microbiol 33:167–176
Gamper HB, Hearst JE (1982) A topological model for transcription based on unwinding angle analysis of E. coli RNA polymerase binary, initiation and ternary complexes. Cell 29:81–90
Hadizadeh Yazdi N, Guet CC, Johnson RC, Marko JF (2012) Variation of the folding and dynamics of the Escherichia coli chromosome with growth conditions. Mol Microbiol 86:1318–1333
Hanawalt PC, Spivak G (2008) Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol 9:958–970
Higgins NP (2014) RNA polymerase: chromosome domain boundary maker and regulator of supercoil density. Curr Opin Microbiol 22C:138–143
Higgins NP, Cozzarelli NR (1982) The binding of gyrase to DNA: analysis by retention to nitrocellulose filters. Nucleic Acids Res 10:6833–6847
Higgins NP, Vologodskii AV (2015) Topological behavior of plasmid DNA. Microbiol Spectr 3(2). doi: 10.1128/microbiolspec.PLAS-0036-2014
Higgins NP, Yang X, Fu Q, Roth JR (1996) Surveying a supercoil domain by using the GD resolution system in Salmonella typhimurium. J Bacteriol 178:2825–2835
Higgins NP, Deng S, Pang Z, Stein R, Champion K, Manna D (2005) Domain behavior and supercoil dynamics in bacterial chromosomes. In: Higgins NP (ed) The bacterial chromosome. ASM Press, Washington, pp 133–153
Ho TD, Figueroa-Bossi N, Wang M, Uzzau S, Bossi L, Slauch JM (2002) Identification of GtgE, a novel virulence factor encoded on the Gifsy-2 bacteriophage of Salmonella enterica serovar Typhimurium. J Bacteriol 184:5234–5239
Holmes VF, Cozzarelli NR (2000) Closing the ring: Links between SMC proteins and chromosome partitioning, condensation, and supercoiling. Proc Natl Acad Sci USA 97:1322–1324
Hutchinson C, Chuang R-Y, Noskov VN et al (2016) Design and synthesis of a minimal bacterial genome. Science. doi: 10.1126/science.aad6253
Johnson RC, Johnson LM, Schmidt JW, Gardner JF (2005) The major nucleoid proteins in the structure and function of the E. coli chromosome. In: Higgins NP (ed) The bacterial chromosome. ASM Press, Washington, pp 65–132
Kirkegaard K, Wang JC (1981) Mapping the topography of DNA wrapped around gyrase by nucleolytic and chemical probing of complexes of unique DNA sequences. Cell 23:721–729
Kleckner N, Fisher JK, Stouf M, White MA, Bates D, Witz G (2014) The bacterial nucleoid: nature, dynamics and sister segregation. Curr Opin Microbiol 22:127–137
Knight SW, Samuels DS (1999) Natural synthesis of a DNA-binding protein from the C-terminal domain of DNA gyrase A in Borrelia burgdorferi. EMBO J 18:4875–4881
Liu LF, Wang JC (1987) Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA 84:7024–7027
Manna D, Wang X, Higgins NP (2001) Mu and Is1 transposition exhibits strong orientation bias at the E. coli bgl locus. J Bacteriol 183:3328–3335
Oram M, Travers AA, Howells AJ, Maxwell A, Pato ML (2006) Dissection of the bacteriophage Mu strong gyrase site (SGS): significance of the SGS right arm in Mu biology and DNA gyrase mechanism. J Bacteriol 188:619–632
Pang Z, Chen R, Manna D, Higgins NP (2005) A gyrase mutant with low activity disrupts supercoiling at the replication terminus. J Bacteriol 187:7773–7783
Pato ML, Banerjee M (2000) Genetic analysis of the strong gyrase site (SGS) of bacteriophage Mu: localization of determinants required for promoting Mu replication. Mol Microbiol 37:800–810
Postow L, Hardy CD, Arsuaga J, Cozzarelli NR (2004) Topological domain structure of the Escherichia coli chromosome. Genes Dev 18:1766–1779
Pul U, Wagner R (2010) Nucleoid-associated proteins: structural properties. In: Dorman CJ, Dame RT (eds) Bacterial chromatin. Springer, New York, pp 175–204
Rovinskiy N, Agbleke AA, Chesnokova O, Pang Z, Higgins NP (2012) Rates of gyrase supercoiling and transcription elongation control supercoil density in a bacterial chromosome. PLoS Genet 8:e1002845
Ruthenburg AJ, Graybosch DM, Huetsch JC, Verdine GL (2005) A superhelical spiral in the Escherichia coli DNA gyrase A C-terminal domain imparts unidirectional supercoiling bias. J Biol Chem 280:26177–26184
Sawitzke JA, Austin S (2000) Suppression of chromosome segregation defects of Escherichia coli muk mutants by mutations in topoisomerase I. Proc Natl Acad Sci USA 97:1671–1676
Scheirer KE, Higgins NP (1997) The DNA cleavage reaction of DNA gyrase. Comparison of stable ternary complexes formed with enoxacin and CcdB protein. J Biol Chem 272:27202–27209
Stein R, Deng S, Higgins NP (2005) Measuring chromosome dynamics on different timescales using resolvases with varying half-lives. Mol Microbiol 56:1049–1061
Stern MJ, Ames GF-L, Smith M, Robinson EC, Higgins CF (1984) Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell 37:1015–1026
Tehranchi AK, Blankschien MD, Zhang Y, Halliday JA, Srivatsan A, Peng J, Herman C, Wang JD (2010) The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell 141:595–605
Tretter EM, Berger JM (2012a) Mechanisms for defining supercoiling set point of DNA gyrase orthologs: I. A nonconserved acidic C-terminal tail modulates Escherichia coli gyrase activity. J Biol Chem 287:18636–18644
Tretter EM, Berger JM (2012b) Mechanisms for defining supercoiling set point of DNA gyrase orthologs: II. The shape of the GyrA subunit C-terminal domain (CTD) is not a sole determinant for controlling supercoiling efficiency. J Biol Chem 287:18645–18654
Wang X, Reyes-Lamothe R, Sherratt DJ (2008) Modulation of Escherichia coli sister chromosome cohesion by topoisomerase IV. Genes Dev 22:2426–2433
Wendorff TJ, Schmidt BH, Heslop P, Austin CA, Berger JM (2012) The structure of DNA-bound human topoisomerase II alpha: conformational mechanisms for coordinating inter-subunit interactions with DNA cleavage. J Mol Biol 424:109–124
Wiggins PA, Cheveralls KC, Martin JS, Lintner R, Kondev J (2010) Strong intranucleoid interactions organize the Escherichia coli chromosome into a nucleoid filament. Proc Natl Acad Sci USA 107:4991–4995
Yang Y, Ames GF (1988) DNA gyrase binds to the family of prokaryotic repetitive extragenic palindromic sequences. Proc Natl Acad Sci USA 85:8850–8854
Acknowledgments
Work in the Higgins lab has been supported by the United States National Institutes of Health grant GM-33143.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest statement
The author declares that he has no conflict of interest regarding this article.
Ethical approval statement
This article does not contain any studies with human participants or animals performed by the author.
Additional information
This article is part of a Special Issue on ‘DNA supercoiling, protein interactions and genetic function’ edited by Laura Finzi and Wilma Olson.
Rights and permissions
About this article
Cite this article
Higgins, N.P. Species-specific supercoil dynamics of the bacterial nucleoid. Biophys Rev 8 (Suppl 1), 113–121 (2016). https://doi.org/10.1007/s12551-016-0207-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12551-016-0207-9