Abstract
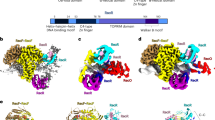
The action of RecA, an important eubacterial protein involved in recombination and repair, involves the transition from an inactive filament in the absence of DNA to an active filament formed in association with DNA and ATP. The structure of the inactive filament was first established in Escherichia coli RecA (EcRecA). The interaction of RecA with non-hydrolysable ATP analogues and ADP has been thoroughly characterized and the DNA binding loops visualized based on the crystal structures of the RecA proteins from Mycobacterium tuberculosis (MtRecA) and Mycobacterium smegmatis (MsRecA). A switch residue, which triggers the transformation of the information on ATP binding to the DNA binding regions, has been identified. The 20-residue C-terminal stretch of RecA, which is disordered in all other relevant crystal structures, has been defined in an MsRecA-dATP complex. The ordering of the stretch is accompanied by the generation of a new nucleotide binding site which can communicate with the original nucleotide binding site of an adjacent molecule in the filament. The plasticity of MsRecA and its mutants involving the switch residue has been explored by studying crystals grown under different conditions at two different temperatures and, in one instance, at low humidity. The structures of these crystals and those of EcRecA and Deinococcus radiodurans RecA (DrRecA) provide information on correlated movements involving different regions of the molecule. These correlated movements appear to be important in the allosteric transitions of RecA during its action.










Similar content being viewed by others
References
Arora A, Chandra NR, Das A, Gopal B, Mande SC, Prakash B, Ramachandran R, Sankaranarayanan R, Sekar K, Suguna K, Tyagi AK, Vijayan M (2011) Structural biology of Mycobacterium tuberculosis proteins: the Indian efforts. Tuberculosis 91(5):456–468. doi:10.1016/j.tube.2011.03.004
Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA 98(18):10037–10041. doi:10.1073/pnas.181342398
Bell CE, Xing X (2004) Crystal structures of Escherichia coli RecA in complex with MgADP and MnAMP-PNP. Biochemistry 43:16142–16152. doi:10.1021/bi048165y
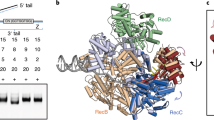
Chen Z, Yang H, Pavletich NP (2008) Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 453:489–494. doi:10.1038/nature06971
Datta S, Prabu MM, Vaze MB, Ganesh N, Chandra NR, Muniyappa K, Vijayan M (2000) Crystal structures of Mycobacterium tuberculosis RecA and its complex with ADP-AlF4: implications for decreased ATPase activity and molecular aggregation. Nucleic Acids Res 28:4964–4973. doi:10.1093/nar/28.24.4964
Datta S, Ganesh N, Chandra NR, Muniyappa K, Vijayan M (2003a) Structural studies on MtRecA-nucleotide complexes: insights into DNA and nucleotide binding and the structural signature of NTP recognition. Proteins 50:474–485. doi:10.1002/prot.10315
Datta S, Krishna R, Ganesh N, Chandra NR, Muniyappa K, Vijayan M (2003b) Crystal structures of Mycobacterium smegmatis RecA and its nucleotide complexes. J Bacteriol 185:4280–4284. doi:10.1128/JB.185.14.4280-4284.2003
Davis EO, Jenner PJ, Brooks PC, Colston MJ, Sedgwick SG (1992) Protein splicing in the maturation of M. tuberculosis RecA protein: a mechanism for tolerating a novel class of intervening sequence. Cell 71:201–210. doi:10.1016/0092-8674(92)90349-H
Dutreix M, Moreau PL, Bailone A, Galibert F, Battista J, Walker GC, Devoret R (1989) New recA mutations that dissociate the various RecA protein activities in Escherichia coli provide evidence for an additional role for RecA protein in UV mutagenesis. J Bacteriol 171:2415–2423
Egelman EH, Stasiak A (1986) Structure of helical RecA-DNA complexes: complexes formed in the presence of ATP-gamma-S or ATP. J Mol Biol 191:677–697. doi:10.1016/0022-2836(86)90453-5
Egelman EH, Stasiak A (1988) Structure of helical RecA-DNA complexes II. Local conformational changes visualized in bundles of RecA-ATPγS filaments. J Mol Biol 200(2):329–349. doi:10.1016/0022-2836(88)90245-8
Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Cryst D66:486–501. doi:10.1107/S0907444910007493
Flory J, Tsang SS, Muniyappa K (1984) Isolation and visualization of active presynaptic filaments of RecA protein and Single-Stranded DNA. Proc Natl Acad Sci USA 81:7026–7030. doi:10.1073/pnas.81.22.7026
Ganesh N, Muniyappa K (2003) Mycobacterium smegmatis RecA protein is structurally similar to but functionally distinct from Mycobacterium tuberculosis RecA. Proteins 53:6–17. doi:10.1002/prot.10433
Kelley JA, Knight KL (1997) Allosteric regulation of RecA protein function is mediated by Gln194. J Biol Chem 272(41):25778–25782. doi:10.1074/jbc.272.41.25778
Krebs WG, Gerstein M (2000) The morph server: a standardized system for analyzing and visualizing macromolecular motions in a database framework. Nucleic Acids Res 28:1665–1675. doi:10.1093/nar/28.8.1665
Krishna R, Manjunath GP, Kumar P, Surolia A, Chandra NR, Muniyappa K, Vijayan M (2006) Crystallographic identification of an ordered C-terminal domain and a second nucleotide-binding site in RecA: new insights into allostery. Nucleic Acids Res 34(8):2186–2195. doi:10.1093/nar/gkl107
Krishna R, Prabu JR, Manjunath GP, Datta S, Chandra NR, Muniyappa K, Vijayan M (2007) Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA. J Mol Biol 367(4):1130–44. doi:10.1016/j.jmb.2007.01.058
Kumar RA, Vaze MB, Chandra NR, Vijayan M, Muniyappa K (1996) Functional characterization of the precursor and spliced forms of RecA protein of Mycobacterium tuberculosis. Biochemistry 35(6):1793–1802. doi:10.1021/bi9517751
Liang JH, Edelsbrunner H, Woodward C (1998) Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design. Protein Sci 7:1884–1897. doi:10.1002/pro.5560070905
Lusetti SL, Shaw JJ, Cox MM (2003a) Magnesium ion-dependent activation of the RecA protein involves the C terminus. J Biol Chem 278:16981–16988. doi:10.1074/jbc.M212916200
Lusetti SL, Wood EA, Fleming CD, Modica MJ, Korth J, Abbott L, Dwyer DW, Roca AI, Inman RB, Cox MM (2003b) C-terminal deletions of the E. coli RecA protein: characterization of in vivo and in vitro effects. J Biol Chem 278:16972–16980. doi:10.1074/jbc.M212917200
Manjunath GP (2009)Mycobacterium smegmatis RecA and SSB: Structure function relationships, interaction with cofactors and accessory proteins. PhD thesis. Indian Institute of Science, Bangalore
McGrew DA, Knight KL (2003) Molecular design and functional organization of the RecA protein. Crit Rev Biochem Mol Biol 38:385–432. doi:10.1080/10409230390242489
Murzin AG, Brenner SE, Hubbard T, Chothia C (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol 247:536–540. doi:10.1016/S0022-2836(05)80134-2
Prabu JR, Manjunath GP, Chandra NR, Muniyappa K, Vijayan M (2008) Functionally important movements in RecA molecules and filaments: studies involving mutation and environmental changes. Acta Crystallographica Section D 64(11):1146–1157. doi:10.1107/S0907444908028448
Rajan R, Bell CE (2004) Crystal structure of RecA from Deinococcus radiodurans: insights into the structural basis of extreme radioresistance. J Mol Biol 344(4):951–963. doi:10.1016/j.jmb.2004.09.087
Story RM, Weber IT, Steitz TA (1992) The structure of the E. coli recA protein monomer and polymer. Nature 355:318–325. doi:10.1038/355318a0
Vaze MB, Muniyappa K (1999) RecA protein from Mycobacterium tuberculosis possesses pH dependent homologous DNA pairing and strand exchange activities: implications for allele exchange in mycobacteria. Biochemistry 38:3175–3186. doi:10.1021/bi9819125
Xing X, Bell CE (2004) Crystal structures of Escherichia coli RecA in a compressed helical filament. J Mol Biol 342:1471–1485. doi:10.1016/j.jmb.2004.07.091
Yu X, Egelman EH (1993) The LexA repressor binds within the deep helical groove of the activated RecA Filament. J Mol Biol 231:29–40. doi:10.1006/jmbi.1993.1254
Acknowledgements
Many collaborators, graduate students and post-doctoral fellows have contributed to the work reviewed here. Their names appear in the appropriate references. The work is supported by the Department of Biotechnology, Government of India. AVC is a CSIR Junior Research Fellow. MV is a DAE Homi Bhabha Professor.
Conflict of interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chandran, A.V., Vijayan, M. Allosteric movements in eubacterial RecA. Biophys Rev 5, 249–258 (2013). https://doi.org/10.1007/s12551-012-0097-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12551-012-0097-4