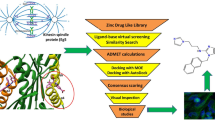
Abstract
Human papillomavirus (HPV), a life-threatening infection, is the leading cause of cancer mortality among women worldwide and needs for designing anticancerous drugs. In the present study, we explored specific novel inhibitors against E6 oncoprotein of high-risk HPV 16, known to inactivate tumor suppressor p53 protein. A homology model of HPV 16 E6 was built and validated using bioinformatics approach. A total of 5000 drug-like compounds were downloaded from ZINC database based on the properties similar to the known inhibitor Jaceosidin (5,7-dihydroxy-2-(4-hydroxy-3-methoxyphenyl)-6-methoxy-4H-chromen-4-one). Virtual-ligand-screening approaches were applied to screen appropriate drug-like compounds using molecular docking program AutoDock Vina in PyRx 0.8, and five best novel drug-like compounds were identified as potential competitive inhibitors against HPV 16 E6 compared to Jaceosidin. Two among these five identified most potential inhibitors, N-[(5-methyl-1H-benzimidazol-2-yl)methyl]-4-oxo-3,4-dihydrophthalazine-1-carboxamide and 6-[3-(3-fluoro-4-methyl-phenyl)-1,2,4-oxadiazol-5-yl]-1,4-dihydroquinoxaline-2,3-dione, were found to interact with E6 with binding energy of \(-7.7\) and \(-7.0\) kcal/mol, respectively, and form H-bonds with p53 binding site of E6 protein residues 113–122 (CQKPLCPEEK). These two inhibitors may help restoration of p53 functioning. The bioinformatics approach extends a promising platform for developing anticancerous competitive inhibitors targeting high-risk HPV 16.



Similar content being viewed by others
References
de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, Plummer M (2012) Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol 13:607–615
Hoory T, Monie A, Gravitt P, Wu TC (2008) Molecular epidemiology of human papillomavirus. J Formos Med Assoc 107:198–217
Parkin DM, Bray F (2006) Chapter 2: the burden of HPV-related cancers. Vaccine 3124:11–25
Guan P, Howell-Jones R, Li N, Bruni L, de Sanjosé S, Franceschi S, Clifford GM (2012) Human papillomavirus types in 115, 789 HPV-positive women: a meta-analysis from cervical infection to cancer. Int J Cancer 131:2349–2359
Chaturvedi AK (2010) Beyond cervical cancer: burden of other HPV-related cancers among men and women. J Adolesc Health 46:20–26
Scheffner M, Munger K, Byrne JC, Howley PM (1991) The state of the p53 and retinoblastoma genes in human cervical carcinoma cell lines. Proc Nat Acad Sci USA 88:5523–5527
Hubbert NL, Sedman SA, Schiller JT (1992) Human papillomavirus type 16 E6 increases the degradation rate of p53 in human keratinocytes. J Virol 66:6237–6241
Talis AL, Huibregtse JM, Howley PM (1998) The role of E6AP in the regulation of p53 protein levels in human papillomavirus (HPV)-positive and HPV-negative cells. J Biol Chem 73:6439–6445
Lee HG, Yu KA, Oh WK, Baeg TW, Oh HC, Ahn JS, Jang WC, Kim JW, Lim JS, Choe YK, Yoon DY (2005) Inhibitory effect of jaceosidin isolated from Artemisia argyi on the function of E6 and E7 oncoproteins of HPV 16. J Ethnopharmacol 98:339–343
Wolf LK (2009) New software and websites for the chemical enterprise. Chem Eng News 87:31
Kelley LA, Sternberg MJ (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371
Krieger E, Joo K, Lee J, Lee J, Raman S, Thompson J, Tyka M, Baker D, Karplus K (2009) Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins 77:114–122
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35:407–410
Wallner B, Elofsson A (2003) Can correct protein models be identified? Protein Sci 12:1073–1086
Eisenberg D, Lüthy R, Bowie JU (1997) VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol 277:396–404
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of non bonded atomic interactions. Protein Sci 2:1511–1519
Irwin JJ, Shoichet BK (2005) ZINC-a free database of commercially available compounds for virtual screening. J Chem Inf Model 45:177–182
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461
Zanier K, Charbonnier S, Sidi AO, McEwen AG, Ferrario MG, Poussin-Courmontagne P, Cura V, Brimer N, Babah KO, Ansari T, Muller I, Stote RH, Cavarelli J, Vande Pol S, Travé G (2013) Structural basis for hijacking of cellular LxxLL motifs by papillomavirus E6 oncoproteins. Science 339:694–698
Crook T, Tidy JA, Vousden KH (1991) Degradation of p53 can be targeted by HPV E6 sequences distinct from those required for p53 binding and trans-activation. Cell 67:547–556
Oprea TI (2002) Virtual screening in lead discovery: a viewpoint. Molecules 7:51–62
Acknowledgments
Authors express gratitude to the Department of Biotechnology, MoS&T, Government of India, for their financial support to Bioinformatics Centre. Authors thank Dr. B.C. Harinath, Director, JBTDRC and Coordinator, Bioinformatics Centre for his insightful comments and suggestions. Grateful thanks to Shri D.S. Mehta, President, Kasturba Health Society, Dr. (Mrs.) P. Narang, Secretary, Kasturba Health Society, Dr. B.S. Garg, Dean, MGIMS and Dr. S.P. Kalantri, Medical Superintendent, Kasturba Hospital Sevagram for their encouragement and unconditional support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kumar, S., Jena, L., Mohod, K. et al. Virtual Screening for Potential Inhibitors of High-Risk Human Papillomavirus 16 E6 Protein. Interdiscip Sci Comput Life Sci 7, 136–142 (2015). https://doi.org/10.1007/s12539-015-0008-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-015-0008-z