Abstract
Adult B-lineage acute lymphoblastic leukemia (B-ALL) with t(4;11)(q21;q23) is very rare. It is characterized by mixed-lineage leukemia and has the potential for lineage switching during the treatment course. We report the disease course of a patient with B-ALL with t(4;11)(q21;q23) to demonstrate that close monitoring of cell morphology and immunophenotyping is necessary to capture the lineage switch at an early stage. Cell morphology, immunophenotyping, and cytogenetics were used to evaluate the patient’s disease status. A 36-year-old woman was diagnosed with B-ALL with t(4;11)(q21;q23), which encodes the KMT2A::AFF1 fusion. After the initial induction chemotherapy, her disease remained refractory, and the patient received salvage immunotherapy with blinatumomab and inotuzumab ozogamicin. However, the ALL did not respond. Repeated bone marrow examinations unexpectedly revealed the emergence of a major population of monoblasts, in addition to a minor population of the original B lymphoblasts. The patient was diagnosed with disease evolution from B-ALL to mixed-phenotype acute leukemia (MPAL, B/myeloid). We present this case to highlight the potential of KMT2A-rearranged B-ALL to undergo lineage switch following B-cell targeted therapy. Patients with this kind of B-ALL should therefore be closely monitored to capture potential changes in the nature of the disease and prompt appropriate treatment.
Similar content being viewed by others
Introduction
Histone-lysine N-methyltransferase 2A (KMT2A), also known as the mixed-lineage leukemia (MLL) gene, is located at 11q23 and has been reported to have more than 100 different types of fusion partners in the literature, with t(4;11)(q21;q23) being the most common [1]. Overall, KMT2A rearrangement can be found in more than 70% of infantile B-lineage acute lymphoblastic leukemia (B-ALL), 5–6% of pediatric case, and about 15% of adult cases [2, 3]. The incidence of t(4;11)(q21;q23) translocation, which leads to the KMT2A::AFF1 fusion gene, in adult B-ALL is approximately 5%; however, its association with unique characteristics in cancer biology and clinical presentation has raised a great deal of interest. Bispecific T-cell engagers (BiTEs) has become an important therapeutic option for B-ALL and usually takes the form of a monoclonal antibody targeting CD19 and CD3 on B and T cells, respectively, causing B lymphoblasts to be directly lysed by cytotoxic T cells. Inotuzumab ozogamicin, an anit-CD22 monoclonal antibody-drug conjugate, is another established treatment option for relapsed and refractory B-ALL. To date, a few cases of KMT2A-rearranged ALL that show a lineage-switch into mixed phenotype acute leukemia (MPAL) after BiTE therapy have been reported [4,5,6]. In this report, we present the case of a 36-year-old woman who was diagnosed with B-ALL with t(4;11)(q21;q23), which transformed into MPAL soon after two lines of B lymphoblast-targeted therapies.
Clinical history
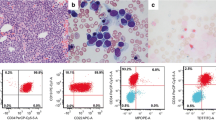
A 36-year-old woman who had been previously healthy visited a local hospital presented with fever and exertional dyspnea. Complete peripheral blood (PB) counts showed pancytopenia with circulating blasts. Bone marrow (BM) examination revealed an extremely hypercellular BM with more than 70% blasts that were positive for CD19 and cytoplasmic CD79a, CD45, and CD22, and negative for CD34 (majority), CD10, CD20, CD117, and CD33 (Fig. 1a, Supplementary Fig. 1). Cytogenetic analysis revealed t(4;11)(q21;q23) in 18 of the 20 analyzed cells (Fig. 1b). The patient then received induction chemotherapy with a hyperCVAD regimen. However, after two courses of chemotherapy, follow-up BM aspiration revealed refractory disease, and the patient was referred to our hospital for further evaluation and treatment, including allogeneic hematopoietic stem cell transplantation.
a Immunophenotyping of the patient’s bone marrow specimen at initial diagnosis. The B-ALL lymphoblasts were CD19+, cyCD79a+, CD34-(majority). Red, CD34+ lymphoblasts; orange, CD34− lymphoblasts; green, granulocytes; pink, residual normal mature B-cells. SSC (side scatter). b Cytogenetic analysis of the patient’s bone marrow at initial diagnosis revealed presence of t(4;11)(q21;q23) (red arrow). Detailed karyotypes: 46,XX,t(4;11)(q21;q23)[13]/45,XX, t(4;11),dic(17;19)(p11.2:p13.2)[2]/47,XX,+X,t(4;11), dic(17;19),+19[3]/46,XX[2]
Material and methods
Cell morphology was evaluated with a bone marrow aspiration smear stained with Liu’s stain (a modified Romanowsky stain). Myeloperoxidase (MPO) and chloroacetate esterase /α-naphthyl butyrate esterase (ANBE) staining were performed to facilitate lineage assignment. Immunophenotyping was performed on bone marrow aspirates using an eight-color assay with internal controls to determine positive and negative patterns. Cytogenetic analysis was performed using a conventional G-banding technique.
Results
After admission, the BOMES regimen (BCNU, vincristine, methotrexate etoposide, and methylprednisolone) was administered for re-induction; however, follow-up BM examination still showed refractory disease with leukemic cells accounting for 72.37% of total nucleated cells (Fig. 2a); all of which expressed CD19 and CD22, but were negative for CD34 (majority), CD10, and CD20 (Fig. 2b). Blinatumomab was administered as a salvage treatment, but the disease persisted. Therefore, inotuzumab ozogamicin was administered because her lymphoblasts tested positive for CD22. However, all treatments failed to induce remission, and elevated PB blasts were still observed one week later. Interestingly, we noticed that the morphology of the PB blasts at this time became atypical for lymphoblasts, with most blasts having a monocytoid appearance with abundant cytoplasm and folded nuclei. Therefore, BM examination was performed again to ascertain the patient’s disease status. The BM smear revealed the presence of two blast populations: a smaller population had a lymphoblast-like morphology and stained negative for MPO and ANBE, whereas a larger blast population exhibited a monoblast-like morphology and negatively stained for MPO but positively stained for ANBE (Fig. 3a). Immunophenotyping using flow cytometry also confirmed that the two blast clones were lymphoblasts, 15.23% (CD34−, CD19+, CD10−, and CD20−) and monoblasts, 67.35% (CD34−, HLA-DR−, partial CD117+, partial CD11b+, CD64+, partial CD14+, CD33+, CD15+, and CD56+). Notably, both clones expressed neuron-glial antigen-2 (NG2), a marker that is strongly associated with KMT2A rearrangement (Fig. 3b). Moreover, cytogenetic examination showed persistent t(4;11)(q21;q23) (Fig. 3c). Therefore, disease evolution from B-ALL to MPAL (B/myeloid) was diagnosed. The ENOP (Etoposide, MitoXANtrone, VinCristine, 6-MP) regimen was administered as salvage therapy. Unfortunately, her disease did not respond either, and the patient quickly succumbed to rapidly progressive leukemia and superimposed infections.
Follow-up bone marrow examination of the patient. a The patient’s marrow was filled with monotonous lymphoblasts with immature chromatin morphology and high N/C ratio (Liu’s stain, 1000×). The myeloperoxidase (MPO) stain was negative. b Flow cytometric immunophenotyping revealed that leukemic cells expressed CD19+, CD22+, CD10−, CD20−, CD34- (majority), CD38+. Red, lymphoblasts; green, granulocytes; blue, mature lymphocytes. SSC (side scatter)
a Morphological examination of the patient’s bone marrow post blinatumomab and inotuzumab ozogamicin treatment. In the Liu’s stain, two populations of abnormal cells are observed, one resembles monoblasts (red arrows), and the other resembles lymphoblasts (black arrows). The monoblasts are myeloperoxidase negative and ANBE positive, while the lymphoblasts are negative for both. b Immunophenotyping of the patient’s bone marrow post blinatumomab and inotuzumab ozogamicin treatment. The lymphoblastic clone (red) are CD19+, CD34−, CD10−, CD38+, HLA-DR+; while the monoblastic clone (light green) are CD19−, CD64+, partial CD14+, partial 11b+, CD33+, CD56+, CD38+, CD15+, HLA-DR−. Both leukemic clones expressed NG2, a biomarker of KMT2A rearrangement. SSC (side scatter). c Follow-up cytogenetics in peripheral blood specimen showed persistent existence of t(4;11) (q21;q23) (red arrow). Detailed karyotypes: 45-52,XX,+der(4)t(4;11)(q21;q23),t(4;11),+dup(7)(q22q32),+8,+12,+13,+19[cp20]
Discussion
B-ALL with t(4;11)(q21;q23) comprises about 5% of adult B-ALL and is categorized as B-lymphoblastic leukemia/lymphoma with KMT2A rearrangement in the updated 5th edition WHO Classification, and as B-ALL with t(v;11q23.3)/KMT2A rearranged in the 2022 International Consensus Classification (ICC) classification [7, 8]. This category of leukemia is characterized by mixed-lineage leukemia and potential for lineage switching during the treatment course. Immunophenotypically, B-ALL with t(4;11) usually expresses CD19, CD34, and TdT, but not CD10 or CD20, which corresponds to very immature B precursors (pro-B cell immunophenotype). Clinically, patients with B-ALL with t(4;11) usually present with leukocytosis and extramedullary involvement (including central nervous system and hepatosplenic) and have a grave prognosis. In terms of molecular pathogenesis, t(4;11) (q21;q23) translocation results in KMT2A::AFF1 fusion (previously known as MLL-AF4), which causes aberrant overexpression of a number of genes, including the anti-apoptotic BCL2 and the proto-oncogene MYC, ultimately driving leukemogenesis [9].
The lineage switch is a unique phenomenon that may occur in B-ALL. Although KMT2A-rearranged B-ALL is the most commonly documented disease that has the potential of lineage switch, ZNF384 translocations, especially TCF3-ZNF384 or TAF15-ZNF384 had also been reported to be implicated in lineage switch process [10, 11].
According to the literature, the lineage switch from B-ALL with KMT2A::AFF1 fusion is usually bi-lineal, with the myeloid component most commonly being monocytic [12,13,14]. However, the exact mechanisms leading to the lineage switch remain unclear. B-ALL with KMT2A::AFF1 fusion has unique characteristics at presentation, including a high WBC count, propensity for CNS involvement, splenomegaly, and poor clinical outcomes. Immunophenotypically, this type of B-ALL is usually CD10− and NG2+ and is classified as Pro-B-ALL [15].
The patient presented in this report was initially diagnosed with B-ALL as evidenced by the typical features on morphological examination and immunophenotyping. However, the patient demonstrated resistance to many lines of B-ALL-directed chemotherapeutic and targeted agents and the B-ALL evolved into MPAL. This case was particularly intriguing because the MPAL transformation occurred soon after two potent targeted therapies, blinatumomab and inotuzumab ozogamicin, were administered. Maybe there was a very small clone of monocytic blasts at initial diagnosis, which became the dominant leukemic clone during disease progression. However, when the patient was transferred to our hospital, the flow cytometry study collected 500,000 events and thus could reached a theoretical limit of sensitivity of 1 × 10−4, so the likelihood of this hypothesis was rather low. Furthermore, we reason that it was more likely that the harsh selection pressure of multiple rounds of B-ALL directed chemotherapy and targeted agents eradicated the bulk of the patient’s initial leukemic burden. However, precursor leukemic cells harboring the KMT2A::AFF1 fusion could theoretically possess lineage plasticity that were later genetically reprogrammed and gave rise to the main monoblastic clone, which lacked CD19 and CD22 expressions, to evade therapeutic killing. In addition, we demonstrated that B lymphoblasts and monoblasts are clonally related, because follow-up cytogenetics of the PB cells showed persistence of t(4;11) (q21;q23), and both populations expressed NG2, as shown using flow cytometry. Therefore, we argue that, for our patient, the lineage switch occurred after B cell-targeted therapy.
Anti-CD19 CAR-T therapy, while being a potent treatment for CD19+ B-ALL, has been reported to induce the lineage switch phenomenon in B-ALL [16]. In a recent study, Jacoby et al. used single-cell technologies to confirm whether leukemic cells with lineage plasticity could undergo cellular reprogramming to switch lineage characteristics following anti-CD19 CAR-T therapy. They further used murine models to demonstrate that a number of key transcription factors, such as PAX5 and ERBF1 are important regulators in this process, because CRISPR/Cas9 mediate ablation of Pax5 or Ebf1 recapitulates the lineage redirection phenomenon under the selection pressure of CD19 CAR [17], BiTEs, such as blinatumomab, efficiently kill CD19+ cancer cells by activating the cytotoxic potential of the patient’s own T cells to lyse tumor cells; thus, the findings presented in the aforementioned study remain relevant in patients with B-ALL undergoing blinatumomab treatment. In the literature, it had been reported in multiple occasions that infantile KMT2A rearranged B-ALL could lineage switch to AML following blinatumomab treatment [6, 18, 19]. In particular, our patient was administered both blinatumomab and inotuzumab ozogamicin, an anti-CD22 antibody-drug conjugate. Therefore, the selection pressure on the B lymphoblast clone was even more intense than that reported in the literature, resulting in the transformation into MPAL with a major monoblastic dominance.
In summary, we present a patient who had been diagnosed with B-ALL, with t(4;11)(q21;q23), treated with multiple lines of chemotherapy and two lines of B cell-directed immunotherapies, and later developed a disease consisting of both B lymphoblasts and monoblasts. We suggest that when managing patients with B-ALL with KMT2A rearrangement, especially the KMT2A::AFF1 fusion, re-evaluation of the nature of leukemic blasts is warranted if the disease becomes refractory to multiple lines of B-ALL-directed therapy. Close monitoring of bone marrow cell morphology and immunophenotyping should be considered at regularly spaced time intervals, for instance every three months, to capture lineage switch at an early stage. Although the prognosis of KMT2A-rearranged B-ALL is dismal, early detection of disease transformation should prompt physicians to consider myeloid leukemia-directed therapy and even stem cell transplantation to successfully salvage these patients.
References
Meyer C, Kowarz E, Hofmann J, Renneville A, Zuna J, Trka J et al (2009) New insights to the MLL recombinome of acute leukemias. Leukemia 23(8):1490–1499. https://doi.org/10.1038/leu.2009.33
Sam TN, Kersey JH, Linabery AM, Johnson KJ, Heerema NA, Hilden JM et al (2012) MLL gene rearrangements in infant leukemia vary with age at diagnosis and selected demographic factors: a Children’s Oncology Group (COG) study. Pediatr Blood Cancer 58(6):836–839. https://doi.org/10.1002/pbc.23274
Forgione MO, McClure BJ, Eadie LN, Yeung DT, White DL (2020) KMT2A rearranged acute lymphoblastic leukaemia: unravelling the genomic complexity and heterogeneity of this high-risk disease. Cancer Lett 469:410–418. https://doi.org/10.1016/j.canlet.2019.11.005
Carulli G, Marini A, Ferreri MI, Azzara A, Ottaviano V, Lari T et al (2012) B-cell acute lymphoblastic leukemia with t(4;11)(q21;q23) in a young woman: evolution into mixed phenotype acute leukemia with additional chromosomal aberrations in the course of therapy. Hematol Rep. 4(3):e15. https://doi.org/10.4081/hr.2012.e15
Haddox CL, Mangaonkar AA, Chen D, Shi M, He R, Oliveira JL et al (2017) Blinatumomab-induced lineage switch of B-ALL with t(4:11)(q21;q23) KMT2A/AFF1 into an aggressive AML: pre- and post-switch phenotypic, cytogenetic and molecular analysis. Blood Cancer J 7(9):e607. https://doi.org/10.1038/bcj.2017.89
Rayes A, McMasters RL, O'Brien MM (2016) Lineage switch in MLL-rearranged infant leukemia following CD19-directed therapy. Pediatr Blood Cancer 63(6):1113–1115. https://doi.org/10.1002/pbc.25953
Arber DA, Orazi A, Hasserjian RP, Borowitz MJ, Calvo KR, Kvasnicka HM et al (2022) International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: integrating morphologic, clinical, and genomic data. Blood 140(11):1200–1228. https://doi.org/10.1182/blood.2022015850
Alaggio R, Amador C, Anagnostopoulos I, Attygalle AD, Araujo IBO, Berti E et al (2022) The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 36(7):1720–1748. https://doi.org/10.1038/s41375-022-01620-2
Harman JR, Thorne R, Jamilly M, Tapia M, Crump NT, Rice S et al (2021) A KMT2A-AFF1 gene regulatory network highlights the role of core transcription factors and reveals the regulatory logic of key downstream target genes. Genome Res. https://doi.org/10.1101/gr.268490.120
Grammatico S, Vitale A, La Starza R, Gorello P, Angelosanto N, Negulici AD et al (2013) Lineage switch from pro-B acute lymphoid leukemia to acute myeloid leukemia in a case with t(12;17)(p13;q11)/TAF15-ZNF384 rearrangement. Leuk Lymphoma 54(8):1802–1805. https://doi.org/10.3109/10428194.2012.753450
Oberley MJ, Gaynon PS, Bhojwani D, Pulsipher MA, Gardner RA, Hiemenz MC et al (2018) Myeloid lineage switch following chimeric antigen receptor T-cell therapy in a patient with TCF3-ZNF384 fusion-positive B-lymphoblastic leukemia. Pediatr Blood Cancer 65(9):e27265. https://doi.org/10.1002/pbc.27265
Dunphy CH, Gardner LJ, Evans HL, Javadi N (2001) CD15(+) acute lymphoblastic leukemia and subsequent monoblastic leukemia: persistence of t(4;11) abnormality and B-cell gene rearrangement. Arch Pathol Lab Med 125(9):1227–1230. https://doi.org/10.5858/2001-125-1227-CALLAS
Nagasaka M, Maeda S, Maeda H, Chen HL, Kita K, Mabuchi O et al (1983) Four cases of t(4;11) acute leukemia and its myelomonocytic nature in infants. Blood 61(6):1174–1181
Stong RC, Korsmeyer SJ, Parkin JL, Arthur DC, Kersey JH (1985) Human acute leukemia cell line with the t(4;11) chromosomal rearrangement exhibits B lineage and monocytic characteristics. Blood 65(1):21–31
Marchesi F, Girardi K, Avvisati G (2011) Pathogenetic, clinical, and prognostic features of adult t(4;11)(q21;q23)/MLL-AF4 positive B-cell acute lymphoblastic leukemia. Adv Hematol 2011:621627. https://doi.org/10.1155/2011/621627
Lucero OM, Parker K, Funk T, Dunlap J, Press R, Gardner RA et al (2019) Phenotype switch in acute lymphoblastic leukaemia associated with 3 years of persistent CAR T cell directed-CD19 selective pressure. Br J Haematol 186(2):333–336. https://doi.org/10.1111/bjh.15812
Jacoby E, Nguyen SM, Fountaine TJ, Welp K, Gryder B, Qin H et al (2016) CD19 CAR immune pressure induces B-precursor acute lymphoblastic leukaemia lineage switch exposing inherent leukaemic plasticity. Nat Commun 7:12320. https://doi.org/10.1038/ncomms12320
Du J, Chisholm KM, Tsuchiya K, Leger K, Lee BM, Rutledge JC et al (2021) Lineage switch in an infant B-lymphoblastic leukemia with t(1;11)(p32;q23); KMT2A/EPS15, following blinatumomab therapy. Pediatr Dev Pathol 24(4):378–382. https://doi.org/10.1177/10935266211001308
Semchenkova A, Mikhailova E, Komkov A, Gaskova M, Abasov R, Matveev E et al (2022) Lineage conversion in pediatric B-cell precursor acute leukemia under blinatumomab therapy. Int J Mol Sci 23(7). https://doi.org/10.3390/ijms23074019
Acknowledgements
The authors thank Division of Hematology of Department of Internal Medicine and Department of Laboratory Medicine, National Taiwan University Hospital for providing accurate diagnostic procedures and joint patient care.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was not supported by any funding.
Conflicts of interest
The authors declare no competing interests.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, JR., Shih, PC., Li, C. et al. Lineage switch of KMT2A-rearranged adult B-lineage acute lymphoblastic leukemia following bispecific T-cell engager and monoclonal antibody therapy. J Hematopathol 16, 103–109 (2023). https://doi.org/10.1007/s12308-023-00539-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12308-023-00539-6