Abstract
Background
Metastasis and drug resistance remain a persistent key clinical obstacle to the success of breast cancer treatments. Recent years have seen an increased focus on understanding the factors that influence metastasis and drug resistance.
Methods
In this study, the changes in MMPs gene expression were investigated together with their regulatory pathways—PI3K, MAPK and NFKβ pathways—during the process of developing tamoxifen resistance in MCF7 cell line. Gene correlation maps and Kaplan–Meier survival plots among all breast cancer patients and patients treated with tamoxifen were evaluated.
Results
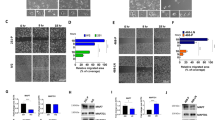
MMPs gene expression was found to be up regulated in MCF7 cell line treated with tamoxifen during the development of tamoxifen resistance using two approaches. Up-regulation of gene expression of AKT1 and MAPK1 started in cells treated with 10 μM tamoxifen that was followed with up-regulation of other genes in these pathways and MMPs in cells treated with 35 μM tamoxifen. MMPs and genes from PI3K, MAPK and NFKβ pathways showed highly significant increase of expression at 50 μM or when cells were treated sequentially six times with 35 μM. Furthermore, increased genes expression was associated with aggressive pattern, clear morphological changes, higher growth rate, increased migration and adhesion potential and tamoxifen insensitivity. Breast cancer distant metastasis-free survival, and survival among tamoxifen treated patients had high expression levels of MAPK1, AKT1, TIMP2, MMP1, and MMP9 showed poor prognosis.
Conclusion
Early changes of MAPK1, AKT1 gene expression upon tamoxifen treatment could possibly be used as an early marker of resistance and future poor prognosis.








Similar content being viewed by others
References
Makki J. Diversity of breast carcinoma: histological subtypes and clinical relevance. Clin Med Insights Pathol. 2015;8:23–31. https://doi.org/10.4137/CPath.S31563.
Waks AG, Winer EP. Breast cancer treatment: a review. JAMA. 2019;321:288–300.
Eliyatkın N, Yalçın E, Zengel B, Aktaş S, Vardar E. Molecular classification of breast carcinoma: from traditional, old-fashioned way to a new age, and a new way. J Breast Health. 2015;11:59–66.
Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. JNCI J Natl Cancer Inst. 2009;101:736–50.
Patel HK, Bihani T. Selective estrogen receptor modulators (SERMs) and selective estrogen receptor degraders (SERDs) in cancer treatment. Pharmacol Ther. 2018;186:1–24.
Araki K, Miyoshi Y. Mechanism of resistance to endocrine therapy in breast cancer: the important role of PI3K/Akt/mTOR in estrogen receptor-positive, HER2-negative breast cancer. Breast Cancer. 2018;5:392–401.
AlFakeeh A, Brezden-Masley C. Overcoming endocrine resistance in hormone receptor–positive breast cancer. Curr Oncol. 2018;25(Suppl 1):S18–27.
Wilson S, Chia SK. Treatment algorithms for hormone receptor-positive advanced breast cancer: applying the results from recent clinical trials into daily practice—insights, limitations, and moving forward. Am Soc Clin Oncol Educ Book. 2013;33:e20–7.
Haque M, Desai KV. Pathways to endocrine therapy resistance in breast cancer. Front Endocrinol. 2019;10:573.
Heiney SP, Parker PD, Felder TM, Adams SA, Omofuma OO, Hulett JM. A systematic review of interventions to improve adherence to endocrine therapy. Breast Cancer Res Treat. 2019;173:499–510.
Hamadneh L, Abuarqoub R, Alhusban A, Bahader M. Upregulation of PI3K/AKT/PTEN pathway is correlated with glucose and glutamine metabolic dysfunction during tamoxifen resistance development in MCF-7 cells. Sci Rep. 2020;10(1):1–7. https://doi.org/10.1038/s41598-020-78833-x.
Kastrati I, Joosten SE, Semina SE, Alejo LH, Brovkovych SD, Stender JD, et al. The NFκB pathway promotes tamoxifen tolerance and disease recurrence in estrogen receptor positive breast cancers. Mol Cancer Res. 2020;18:1018–27.
Xu W, Yang Z, Lu N. A new role for the PI3K/Akt signaling pathway in the epithelial-mesenchymal transition. Cell Adh Migr. 2015;9:317–24.
Roy R, Morad G, Jedinak A, Moses MA. Metalloproteinases and their roles in human cancer. Anat Rec. 2020;303:1557–72.
Abedin M, King N. Diverse evolutionary paths to cell adhesion. Trends Cell Biol. 2010;20:734–42.
Wang Q, Gun M, Hong XY. Induced tamoxifen resistance is mediated by increased methylation of e-cadherin in estrogen receptor-expressing breast cancer cells. Sci Rep. 2019;9:1–7.
Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res Treat. 2010;123:725–31.
Shen CJ, Kuo YL, Chen CC, Chen MJ, Cheng YM. MMP1 expression is activated by Slug and enhances multi-drug resistance (MDR) in breast cancer. PLoS ONE. 2017;12:e0174487.
Jiang Q, Pan Y, Cheng Y, Li H, Liu D, Li H. Lunasin suppresses the migration and invasion of breast cancer cells by inhibiting matrix metalloproteinase-2/-9 via the FAK/Akt/ERK and NF-κB signaling pathways. Oncol Rep. 2016;36:253–62.
Chung TW, Choi H, Lee JM, Ha SH, Kwak CH, Abekura F, et al. Oldenlandia diffusa suppresses metastatic potential through inhibiting matrix metalloproteinase-9 and intercellular adhesion molecule-1 expression via p38 and ERK1/2 MAPK pathways and induces apoptosis in human breast cancer MCF-7 cells. J Ethnopharmacol. 2017;195:309–17.
Zhuang S, Li L, Zang Y, Li G, Wang F. RRM2 elicits the metastatic potential of breast cancer cells by regulating cell invasion, migration and VEGF expression via the PI3K/AKT signaling. Oncol Lett. 2020;19:3349–55.
Yang H, Liang J, Zhou J, Mi J, Ma K, Fan Y, et al. Knockdown of RHOC by shRNA suppresses invasion and migration of cholangiocellular carcinoma cells via inhibition of MMP2, MMP3, MMP9 and epithelial-mesenchymal transition. Mol Med Rep. 2016;13:5255–61.
Joseph C, Alsaleem M, Orah N, Narasimha PL, Miligy IM, Kurozumi S, et al. Elevated MMP9 expression in breast cancer is a predictor of shorter patient survival. Breast Cancer Res Treat. 2020;182:267–82.
Wang QM, Lv L, Tang Y, Zhang L, Wang LF. MMP-1 is overexpressed in triple-negative breast cancer tissues and the knockdown of MMP-1 expression inhibits tumor cell malignant behaviors in vitro. Oncol lett. 2019;17:1732–40.
Li H, Qiu Z, Li F, Wang C. The relationship between MMP-2 and MMP-9 expression levels with breast cancer incidence and prognosis. Oncol lett. 2017;14:5865–70.
Arpino V, Brock M, Gill SE. The role of TIMPs in regulation of extracellular matrix proteolysis. Matrix Biol. 2015;44:247–54.
Jackson HW, Defamie V, Waterhouse P, Khokha R. TIMPs: versatile extracellular regulators in cancer. Nat Rev Cancer. 2017;17:38–53.
Qin L, Wang Y, Yang N, Zhang Y, Zhao T, Wu Y, et al. Tissue inhibitor of metalloproteinase-1 (TIMP-1) as a prognostic biomarker in gastrointestinal cancer: a meta-analysis. PeerJ. 2021;9:e10859.
Hekmat O, Munk S, Fogh L, Yadav R, Francavilla C, Horn H, et al. TIMP-1 increases expression and phosphorylation of proteins associated with drug resistance in breast cancer cells. J Proteome Res. 2013;12:4136–51.
Remacle A, McCarthy K, Noël A, Maguire T, McDermott E, Ohiggins N, et al. High levels of TIMP-2 correlate with adverse prognosis in breast cancer. Int J Cancer. 2000;89(2):118–21.
Acknowledgements
This project was funded by Al-Zaytoonah University of Jordan research funds (2019–2018/18/06) and (2020–2019/23/06).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
About this article
Cite this article
Hamadneh, L., Bahader, M., Abuarqoub, R. et al. PI3K/AKT and MAPK1 molecular changes preceding matrix metallopeptidases overexpression during tamoxifen-resistance development are correlated to poor prognosis in breast cancer patients. Breast Cancer 28, 1358–1366 (2021). https://doi.org/10.1007/s12282-021-01277-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12282-021-01277-2