Abstract
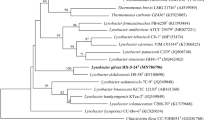
Two Gram-stain-negative, mesophilic, strictly aerobic, nonspore forming, and yellow-pigmented strains with rod-shaped cells, designated H21R20T and H23M41T, were isolated from the faeces of an Oriental stork (Ciconia boyciana). Based on 16S rRNA gene sequences, both strains showed the highest similarity (98.3–98.4%) to the type strain of Lysobacter concretionis. Phylogenetic analysis based on the 16S rRNA genes and 92 bacterial core genes showed that strains H21R20T and H23M41T were robustly clustered with L. concretionis Ko07T. Whole genome sequencing revealed that the genomes of both strains were approximately 2.9 Mb in size. The DNA G + C contents of the H21R20T and H23M41T strains were 67.3 and 66.6%, respectively. The two strains showed 80.1–81.7% average nucleotide identity with L. concretionis Ko07T. Strain H21R20T grew optimally at 30°C and pH 8.0 and in the presence of 0.5–3% (wt/vol) NaCl, while strain H23M41T grew optimally at 30°C and pH 7.0–8.0 and in the presence of 0–3% (wt/vol) NaCl. Both strains possessed iso-C15:0, iso-C16:0 and summed feature 9 (iso-C17:1ω9c and/or C16:010-methyl) as the major cellular fatty acids, ubiquinone Q-8 as a predominant quinone, and diphosphatidylglycerol, phosphatidylglycerol, and phosphatidylethanolamine as the major polar lipids. A multifaceted investigation demonstrated that strains H21R20T and H23M41T represent novel species of the genus Lysobacter, for which we propose the names Lysobacter ciconiae sp. nov. and Lysobacter avium sp. nov. for strains H21R20T (= KCTC 82316T = JCM 34832T) and H23M41T (= KCTC 62676T = JCM 33223T), respectively.
Similar content being viewed by others
References
Arkin, A.P., Cottingham, R.W., Henry, C.S., Harris, N.L., Stevens, R.L., Maslov, S., Dehal, P., Ware, D., Perez, F., Canon, S., et al. 2018. KBase: the United States department of energy systems biology knowledgebase. Nat. Biotechnol. 36, 566–569.
Brettin, T., Davis, J.J., Disz, T., Edwards, R.A., Gerdes, S., Olsen, G.J., Olson, R., Overbeek, R., Parrello, B., Pusch, G.D., et al. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5, 8365.
Chen, I.M.A., Chu, K., Palaniappan, K., Pillay, M., Ratner, A., Huang, J., Huntemann, M., Varghese, N., White, J.R., Seshadri, R., et al. 2019. IMG/M v. 5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 47, D666–D677.
Chhetri, G., Kim, J., Kim, I., and Seo, T. 2019. Lysobacter caseinilyticus, sp. nov., a casein hydrolyzing bacterium isolated from sea water. Antonie van Leeuwenhoek 112, 1349–1356.
Chin, C.S., Alexander, D.H., Marks, P., Klammer, A.A., Drake, J., Heiner, C., Clum, A., Copeland, A., Huddleston, J., Eichler, E.E., et al. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569.
Choi, H., Im, W.T., and Park, J.S. 2018. Lysobacter spongiae sp. nov., isolated from spongin. J. Microbiol. 56, 97–103.
Christensen, P. and Cook, F.D. 1978. Lysobacter, a new genus of nonfruiting, gliding bacteria with a high base ratio. Int. J. Syst. Evol. Microbiol. 28, 367–393.
Chun, J., Oren, A., Ventosa, A., Christensen, H., Arahal, D.R., da Costa, M.S., Rooney, A.P., Yi, H., Xu, X.W., De Meyer, S., et al. 2018. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68, 461–466.
Collins, M.D. and Jones, D. 1981. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol. Rev. 45, 316–354.
Du, J., Singh, H., Ngo, H.T.T., Won, K.H., Kim, K.Y., and Yi, T.H. 2015. Lysobacter tyrosinelyticus sp. nov. isolated from Gyeryongsan national park soil. J. Microbiol. 53, 365–370.
Fang, B.Z., Xie, Y.G., Zhou, X.K., Zhang, X.T., Liu, L., Jiao, J.Y., Xiao, M., and Li, W.J. 2020. Lysobacter prati sp. nov., isolated from a plateau meadow sample. Antonie van Leeuwenhoek 113, 763–772.
Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376.
Ha, S.M., Kim, C.K., Roh, J., Byun, J.H., Yang, S.J., Choi, S.B., Chun, J., and Yong, D. 2019. Application of the whole genome-based bacterial identification system, TrueBac ID, using clinical isolates that were not identified with three matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDITOF MS) systems. Ann. Lab. Med. 39, 530–536.
Hiraishi, A., Ueda, Y., Ishihara, J., and Mori, T. 1996. Comparative lipoquinone analysis of influent sewage and activated sludge by high-performance liquid chromatography and photodiode array detection. J. Gen. Appl. Microbiol. 42, 457–469.
Huo, Y., Kang, J.P., Hurh, J., Han, Y., Ahn, J.C., Mathiyalagan, R., Piao, C., and Yang, D.C. 2018. Lysobacter panacihumi sp. nov., isolated from ginseng cultivated soil. J. Microbiol. 56, 748–752.
Hyun, D.W., Sung, H., Kim, P.S., Yun, J.H., and Bae, J.W. 2021. Leucobacter coleopterorum sp. nov., Leucobacter insecticola sp. nov., and Leucobacter viscericola sp. nov., isolated from the intestine of the diving beetles, Cybister brevis and Cybister lewisianus, and emended description of the genus Leucobacter. J. Microbiol. 59, 360–368.
Im, W.T., Siddiqi, M.Z., Kim, S.Y., Huq, M.A., Lee, J.H., and Choi, K.D. 2020. Lysobacter lacus sp. nov., isolated from from lake sediment. Int. J. Syst. Evol. Microbiol. 70, 2211–2216.
Jang, J.H., Lee, D., and Seo, T. 2018. Lysobacter pedocola sp. nov., a novel species isolated from Korean soil. J. Microbiol. 56, 387–392.
Jeong, S.E., Lee, H.J., and Jeon, C.O. 2016. Lysobacter aestuarii sp. nov., isolated from estuary sediment. Int. J. Syst. Evol. Microbiol. 66, 1346–1351.
Kim, S.J., Ahn, J.H., Weon, H.Y., Hong, S.B., Seok, S.J., Kim, J.S., and Kwon, S.W. 2016. Lysobacter terricola sp. nov., isolated from greenhouse soil. Int. J. Syst. Evol. Microbiol. 66, 1401–1406.
Kim, S.J., Ahn, J.H., Weon, H.Y., Joa, J.H., Hong, S.B., Seok, S.J., Kim, J.S., and Kwon, S.W. 2017. Lysobacter solanacearum sp. nov., isolated from rhizosphere of tomato. Int. J. Syst. Evol. Microbiol. 67, 1102–1106.
Kim, I., Choi, J., Chhetri, G., and Seo, T. 2019. Lysobacter helvus sp. nov. and Lysobacter xanthus sp. nov., isolated from soil in South Korea. Antonie van Leeuwenhoek 112, 1253–1262.
Kim, K.R., Kim, K.H., Khan, S.A., Kim, H.M., Han, D.M., and Jeon, C.O. 2021. Lysobacter arenosi sp. nov. and Lysobacter solisilvae sp. nov. isolated from soil. J. Microbiol. 59, 709–717.
Kluge, A.G. and Farris, J.S. 1969. Quantitative phyletics and the evolution of anurans. Syst. Biol. 18, 1–32.
Kumar, S., Stecher, G., and Tamura, K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874.
Lane, D. 1991. 16S/23S rRNA sequencing. In Stackebrandt, E. and Goodfellow, M. (eds.), Nucleic Acid Techniques in Bacterial Systematics. pp. 115–175. John Wiley and Sons, New York, USA.
Lee, D., Jang, J.H., Cha, S., and Seo, T. 2017. Lysobacter humi sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 67, 951–955.
Lee, S.Y., Kang, W., Kim, P.S., Kim, H.S., Sung, H., Shin, N.R., Whon, T.W., Yun, J.H., Lee, J.Y., Lee, J.Y., et al. 2019. Undibacterium piscinae sp. nov., isolated from Korean shiner intestine. Int. J. Syst. Evol. Microbiol. 69, 3148–3154.
Lee, S.Y., Kang, W., Kim, P.S., Kim, H.S., Sung, H., Shin, N.R., Yun, J.H., Lee, J.Y., Lee, J.Y., Jung, M.J., et al. 2020. Jeotgalibaca ciconiae sp. nov., isolated from the faeces of an Oriental stork. Int. J. Syst. Evol. Microbiol. 70, 3247–3254.
Lee, I., Kim, Y.O., Park, S.C., and Chun, J. 2016. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103.
Lee, S.Y., Sung, H., Kim, P.S., Kim, H.S., Lee, J.Y., Lee, J.Y., Jeong, Y.S., Tak, E.J., Han, J.E., Hyun, D.W., et al. 2021. Description of Ornithinimicrobium ciconiae sp. nov., and Ornithinimicrobium avium sp. nov., isolated from the faeces of the endangered and near-threatened birds. J. Microbiol. 59, 978–987.
Li, W., Elderiny, N.S., Ten, L.N., Lee, S.Y., Kim, M.K., and Jung, H.Y. 2020. Lysobacter terrigena sp. nov., isolated from a Korean soil sample. Arch. Microbiol. 202, 637–643.
Li, L., Stoeckert, C.J.Jr, and Roos, D.S. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189.
Luo, Y., Dong, H., Zhou, M., Huang, Y., Zhang, H., He, W., Sheng, H., and An, L. 2019. Lysobacter psychrotolerans sp. nov., isolated from soil in the Tianshan Mountains, Xinjiang, China. Int. J. Syst. Evol. Microbiol. 69, 926–931.
Margesin, R., Zhang, D.C., Albuquerque, L., Froufe, H.J., Egas, C., and da Costa, M.S. 2018. Lysobacter silvestris sp. nov., isolated from alpine forest soil, and reclassification of Luteimonas tolerans as Lysobacter tolerans comb. nov. Int. J. Syst. Evol. Microbiol. 68, 1571–1577.
Meier-Kolthoff, J.P., Auch, A.F., Klenk, H.P., and Göker, M. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14, 60.
Minnikin, D.E., O’donnell, A.G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., and Parlett, J.H. 1984. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241.
Na, S.I., Kim, Y.O., Yoon, S.H., Ha, S.M., Baek, I., and Chun, J. 2018. UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 56, 280–285.
Parte, A.C., Carbasse, J.S., Meier-Kolthoff, J.P., Reimer, L.C., and Göker, M. 2020. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 70, 5607–5612.
Price, M.N., Dehal, P.S., and Arkin, A.P. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650.
Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sasser, M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. MIDI Inc., Newark, Delaware, USA.
Schaeffer, A.B. and Fulton, M.D. 1933. A simplified method of staining endospores. Science 77, 194.
Siddiqi, M.Z. and Im, W.T. 2016. Lysobacter hankyongensis sp. nov., isolated from activated sludge and Lysobacter sediminicola sp. nov., isolated from freshwater sediment. Int. J. Syst. Evol. Microbiol. 66, 212–218.
Singh, H., Du, J., Ngo, H.T.T., Won, K.H., Yang, J.E., Kim, K.Y., and Yi, T.H. 2015a. Lysobacter fragariae sp. nov. and Lysobacter rhizosphaerae sp. nov. isolated from rhizosphere of strawberry plant. Antonie van Leeuwenhoek 107, 1437–1444.
Singh, H., Du, J., Won, K.H., Yang, J.E., Akter, S., Kim, K.Y., and Yi, T.H. 2015b. Lysobacter novalis sp. nov., isolated from fallow farmland soil. Int. J. Syst. Evol. Microbiol. 65, 3131–3136.
Srinivasan, S., Kim, M.K., Sathiyaraj, G., Kim, H.B., Kim, Y.J., and Yang, D.C. 2010. Lysobacter soli sp. nov., isolated from soil of a ginseng field. Int. J. Syst. Evol. Microbiol. 60, 1543–1547.
Ten, L.N., Jeon, J., Elderiny, N.S., Kim, M.K., Lee, S.Y., and Jung, H.Y. 2020. Lysobacter segetis sp. nov., isolated from soil. Curr. Microbiol. 77, 166–172.
Ten, L.N., Jung, H.M., Im, W.T., Yoo, S.A., and Lee, S.T. 2008. Lysobacter daecheongensis sp. nov., isolated from sediment of stream near the Daechung dam in South Korea. J. Microbiol. 46, 519–524.
Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680.
Xiao, M., Zhou, X.K., Chen, X., Duan, Y.Q., Alkhalifah, D.H.M., Im, W.T., Hozzein, W.N., Chen, W., and Li, W.J. 2019. Lysobacter tabacisoli sp. nov., isolated from rhizosphere soil of Nicotiana tabacum L. Int. J. Syst. Evol. Microbiol. 69, 1875–1880.
Xu, J., Sheng, M., Yang, Z., Qiu, J., Zhang, J., Zhang, L., and He, J. 2021. Lysobacter gilvus sp. nov., isolated from activated sludge. Arch. Microbiol. 203, 7–11.
Ye, X.M., Chu, C.W., Shi, C., Zhu, J.C., He, Q., and He, J. 2015. Lysobacter caeni sp. nov., isolated from the sludge of a pesticide manufacturing factory. Int. J. Syst. Evol. Microbiol. 65, 845–850.
Zhang, L., Bai, J., Wang, Y., Wu, G.L., Dai, J., and Fang, C.X. 2011. Lysobacter korlensis sp. nov. and Lysobacter bugurensis sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 61, 2259–2265.
Zhang, X.J., Yao, Q., Wang, Y.H., Yang, S.Z., Feng, G.D., and Zhu, H.H. 2019. Lysobacter silvisoli sp. nov., isolated from forest soil. Int. J. Syst. Evol. Microbiol. 69, 93–98.
Zhang, H., Yohe, T., Huang, L., Entwistle, S., Wu, P., Yang, Z., Busk, P.K., Xu, Y., and Yin, Y. 2018. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 46, W95–W101.
Acknowledgements
This work was supported by grants from the Mid-Career Researcher Program (NRF-2020R1A2C3012797) through the National Research Foundation of Korea (NRF) and the National Institute of Biological Resources (NIBR201801106) funded by the Ministry of Environment of Korea (MOE). This work was also supported by the NRF grant funded by the Korean government (MSIT) (No. NRF-2018R1A5A1025077).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of Interest The authors declare that there are no conflicts of interest.
Ethical Statement All sampling conducted in this study was approved by the Institutional Animal Care and Use Committee of Kyung Hee University (Permit number: KHUASP(SE)-18-048) and complied with the guidelines of the Committee.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Lee, SY., Kim, P.S., Sung, H. et al. Lysobacter ciconiae sp. nov., and Lysobacter avium sp. nov., isolated from the faeces of an Oriental stork. J Microbiol. 60, 469–477 (2022). https://doi.org/10.1007/s12275-022-1647-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-022-1647-5