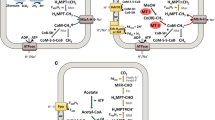
Abstract
Aerobic methane oxidation is a key process in the global carbon cycle that acts as a major sink of methane. In this study, we describe a novel methanotroph designated EMGL16-1 that was isolated from a freshwater lake using the floating filter culture technique. Based on a phylogenetic analysis of 16S rRNA gene sequences, the isolate was found to be closely related to the genus Methylomonas in the family Methylococcaceae of the class Gammaproteobacteria with 94.2–97.4% 16S rRNA gene similarity to Methylomonas type strains. Comparison of chemotaxonomic and physiological properties further suggested that strain EMGL16-1 was taxonomically distinct from other species in the genus Methylomonas. The isolate was versatile in utilizing nitrogen sources such as molecular nitrogen, nitrate, nitrite, urea, and ammonium. The genes coding for subunit of the particulate form methane monooxygenase (pmoA), soluble methane monooxygenase (mmoX), and methanol dehydrogenase (mxaF) were detected in strain EMGL16-1. Phylogenetic analysis of mmoX indicated that mmoX of strain EMGL16-1 is distinct from those of other strains in the genus Methylomonas. This isolate probably represents a novel species in the genus. Our study provides new insights into the diversity of species in the genus Methylomonas and their environmental adaptations.
Similar content being viewed by others
References
Auman, A.J., Speake, C.C., and Lidstrom, M.E. 2001. nifH sequences and nitrogen fixation in type I and type II methanotrophs. Appl. Environ.Microbiol. 67, 4009–4016.
Bedard, C. and Knowles, R. 1989. Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol. Rev. 53, 68–84.
Boden, R., Cunliffe, M., Scanlan, J., Moussard, H., Kits, K.D., Klotz, M.G., Jetten, M.S., Vuilleumier, S., Han, J., Peters, L., et al. 2011. Complete genome sequence of the aerobic marine methanotroph Methylomonas methanica MC09. J. Bacteriol. 193, 7001–7002.
Bowman, J.P., Sly, L.I., Cox, J.M., and Hayward, A.C. 1990. Methylomonas fodinarum sp. nov. and Methylomonas aurantiaca sp. nov.: Two closely related Type I obligate methanotrophs. Syst. Appl. Microbiol. 13, 279–287.
Bowman, J.P., Sly, L.I., Nichols, P.D., and Hayward, A.C. 1993. Revised taxonomy of the methanotrophs: Description of Methylobacter gen. nov., emendation of Methylococcus, validation of Methylosinus and Methylocystis species, and a proposal that the family Methylococcaceae includes only the group I methanotrophs. Int. J. Syst. Evol. Microbiol. 43, 735–753.
Bussmann, I., Rahalkar, M., and Schink, B. 2006. Cultivation of methanotrophic bacteria in opposing gradients of methane and oxygen. FEMS Microbiol. Ecol. 56, 331–344.
Collins, M.D. and Green, P.N. 1985. Isolation and characterization of a novel coenzyme Q from some methane-oxidizing bacteria. Biochem. Biophys. Res. Commun. 133, 1125–1131.
Costello, A.M. and Lidstrom, M.E. 1999. Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl. Environ. Microbiol. 65, 5066–5074.
Danilova, O.V., Kulichevskaya, I.S., Rozova, O.N., Detkova, E.N., Bodelier, P.L., Trotsenko, Y.A., and Dedysh, S.N. 2013. Methylomonas paludis sp. nov., the first acid-tolerant member of the genus Methylomonas, from an acidic wetland. Int. J. Syst. Evol. Microbiol. 63, 2282–2289.
Danilova, O.V., Suzina, N.E., Van De Kamp, J., Svenning, M.M., Bodrossy, L., and Dedysh, S.N. 2016. A new cell morphotype among methane oxidizers: a spiral-shaped obligately microaerophilic methanotroph from northern low-oxygen environments. ISME J. 10, 2734–2743.
de Bruyn, J.C., Boogerd, F.C., Bos, P., and Kuenen, J.G. 1990. Floating filters, a novel technique for isolation and enumeration of fastidious, acidophilic, iron-oxidizing, autotrophic bacteria. Appl. Environ. Microbiol. 56, 2891–2894.
DiSpirito, A.A., Gulledge, J., Shiemke, A.K., Murrell, J.C., Lidstrom, M.E., and Krema, C.L. 1991. Trichloroethylene oxidation by the membrane-associated methane monooxygenase in type I, type II and type X methanotrophs. Biodegradation 2, 151–164.
Dunfield, P.F., Khmelenina, V.N., Suzina, N.E., Trotsenko, Y.A., and Dedysh, S.N. 2003. Methylocella silvestris sp. nov., a novel methanotroph isolated from an acidic forest cambisol. Int. J. Syst. Evol. Microbiol. 53, 1231–1239.
Dunfield, P.F., Yuryev, A., Senin, P., Smirnova, A.V., Stott, M.B., Hou, S., Ly, B., Saw, J.H., Zhou, Z., Ren, Y., et al. 2007. Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 450, 879–882.
Fujie, K., Hu, H.Y., Tanaka, H., Urano, K., Saitou, K., and Katayama, A. 1998. Analysis of respiratory quinones in soil for characterization of microbiota. Soil Sci. Plant Nutr. 44, 393–404.
Griffiths, R.I., Whiteley, A.S., O’Donnell, A.G., and Bailey, M.J. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA-and rRNAbased microbial community composition. Appl. Environ. Microbiol. 66, 5488–5491.
Hanada, S., Shigematsu, T., Shibuya, K., Eguchi, M., Hasegawa, T., Suda, F., Kamagata, Y., Kanagawa, T., and Kurane, R. 1998. Phylogenetic analysis of trichloroethylene-degrading bacteria newly isolated from soil polluted with this contaminant. J. Ferment. Bioengin. 86, 539–544.
Hanson, R.S. and Hanson, T.E. 1996. Methanotrophic bacteria. Microbiol. Rev. 60, 439–471.
Heylen, K., De Vos, P., and Vekeman, B. 2016. Draft genome sequences of eight obligate methane oxidizers occupying distinct niches based on their nitrogen metabolism. Genome Announc. 4, e00421–16.
Hiraishi, A., Ueda, Y., Ishihara, J., and Mori, T. 1996. Comparative lipoquinone analysis of influent sewage and activated sludge by high-performance liquid chromatography and photodiode array detection. J. Gen. Appl. Microbiol. 42, 457–469.
Hoefman, S., Heylen, K., and De Vos, P. 2014a. Methylomonas lenta sp. nov., a methanotroph isolated from manure and a denitrification tank. Int. J. Syst. Evol. Microbiol. 64, 1210–1217.
Hoefman, S., van der Ha, D., Boon, N., Vandamme, P., De Vos, P., and Heylen, K. 2014b. Niche differentiation in nitrogen metabolism among methanotrophs within an operational taxonomic unit. BMC Microbiol. 14, 83.
Hojberg, O., Binnerup, S.J., and Sorensen, J. 1997. Growth of silicone-immobilized bacteria on polycarbonate membrane filters, a technique to study microcolony formation under anaerobic conditions. Appl. Environ. Microbiol. 63, 2920–2924.
Holmes, A.J., Costello, A., Lidstrom, M.E., and Murrell, J.C. 1995. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 132, 203–208.
Hucker, G.J. 1921. A new modification and application of the Gram stain. J. Bacteriol. 6, 395–397.
Hutchens, E., Radajewski, S., Dumont, M.G., McDonald, I.R., and Murrell, J.C. 2004. Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ. Microbiol. 6, 111–120.
Iguchi, H., Sato, I., Sakakibara, M., Yurimoto, H., and Sakai, Y. 2012. Distribution of methanotrophs in the phyllosphere. Biosci. Biotechnol. Biochem. 76, 1580–1583.
Iguchi, H., Yurimoto, H., and Sakai, Y. 2010. Soluble and particulate methane monooxygenase gene clusters of the type I methanotroph Methylovulum miyakonense HT12. FEMS Microbiol. Lett. 312, 71–76.
Iguchi, H., Yurimoto, H., and Sakai, Y. 2011. Stimulation of methanotrophic growth in cocultures by cobalamin excreted by rhizobia. Appl. Environ. Microbiol. 77, 8509–8515.
Kalyuzhnaya, M.G., Khmelenina, V.N., Kotelnikova, S., Holmquist, L., Pedersen, K., and Trotsenko, Y.A. 1999. Methylomonas scandinavica sp. nov., a new methanotrophic psychrotrophic bacterium isolated from deep igneous rock ground water of Sweden. Syst. Appl. Microbiol. 22, 565–572.
Khalifa, A., Lee, C.G., Ogiso, T., Ueno, C., Dianou, D., Demachi, T., Katayama, A., and Asakawa, S. 2015. Methylomagnum ishizawai gen. nov., sp. nov., a mesophilic type I methanotroph isolated from rice rhizosphere. Int. J. Syst. Evol. Microbiol. 65, 3527–3534.
Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120.
Kits, K.D., Campbell, D.J., Rosana, A.R., and Stein, L.Y. 2015a. Diverse electron sources support denitrification under hypoxia in the obligate methanotroph Methylomicrobium album strain BG8. Front. Microbiol. 6, 1072.
Kits, K.D., Klotz, M.G., and Stein, L.Y. 2015b. Methane oxidation coupled to nitrate reduction under hypoxia by the Gammaproteobacterium Methylomonas denitrificans, sp. nov. type strain FJG1. Environ. Microbiol. 17, 3219–3232.
Knief, C. 2015. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front. Microbiol. 6, 1346.
Koh, S.C., Bowman, J.P., and Sayler, G.S. 1993. Soluble methane monooxygenase production and trichloroethylene degradation by a Type I methanotroph, Methylomonas methanica 68-1. Appl. Environ. Microbiol. 59, 960–967.
Kolb, S., Knief, C., Stubner, S., and Conrad, R. 2003. Quantitative detection of methanotrophs in soil by novel pmoA-targeted realtime PCR assays. Appl. Environ. Microbiol. 69, 2423–2429.
Kumar, S., Stecher, G., and Tamura, K. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874.
Lee, S.W., Im, J., Dispirito, A.A., Bodrossy, L., Barcelona, M.J., and Semrau, J.D. 2009. Effect of nutrient and selective inhibitor amendments on methane oxidation, nitrous oxide production, and key gene presence and expression in landfill cover soils: characterization of the role of methanotrophs, nitrifiers, and denitrifiers. Appl. Microbiol. Biotechnol. 85, 389–403.
McDonald, I.R., Kenna, E.M., and Murrell, J.C. 1995. Detection of methanotrophic bacteria in environmental samples with the PCR. Appl. Environ. Microbiol. 61, 116–121.
McDonald, I.R. and Murrell, J.C. 1997. The methanol dehydrogenase structural gene mxaF and its use as a functional gene probe for methanotrophs and methylotrophs. Appl. Environ. Microbiol. 63, 3218–3224.
Morton, J.D., Hayes, K.F., and Semrau, J.D. 2000. Effect of copper speciation on whole-cell soluble methane monooxygenase activity in Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 66, 1730–1733.
Nei, M., Kumar, S., and Takahashi, K. 1998. The optimization principle in phylogenetic analysis tends to give incorrect topologies when the number of nucleotides or amino acids used is small. Proc. Natl. Acad. Sci. USA 95, 12390–12397.
Nyerges, G. and Stein, L.Y. 2009. Ammonia cometabolism and product inhibition vary considerably among species of methanotrophic bacteria. FEMS Microbiol. Lett. 297, 131–136.
Ogiso, T., Ueno, C., Dianou, D., Huy, T.V., Katayama, A., Kimura, M., and Asakawa, S. 2012. Methylomonas koyamae sp. nov., a type I methane-oxidizing bacterium from floodwater of a rice paddy field. Int. J. Syst. Evol. Microbiol. 62, 1832–1837.
Oldenhuis, R., Oedzes, J.Y., van der Waarde, J.J., and Janssen, D.B. 1991. Kinetics of chlorinated hydrocarbon degradation by Methylosinus trichosporium OB3b and toxicity of trichloroethylene. Appl. Environ. Microbiol. 57, 7–14.
Oswald, K., Graf, J.S., Littmann, S., Tienken, D., Brand, A., Wehrli, B., Albertsen, M., Daims, H., Wagner, M., Kuypers, M.M., et al. 2017. Crenothrix are major methane consumers in stratified lakes. ISME J. 2017, 1–17.
Park, S., Shah, N.N., Taylor, R.T., and Droege, M.W. 1992. Batch cultivation of Methylosinus trichosporium OB3b: II. Production of particulate methane monooxygenase. Biotechnol. Bioeng. 40, 151–157.
Pol, A., Heijmans, K., Harhangi, H.R., Tedesco, D., Jetten, M.S., and Opden Camp, H.J. 2007. Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature 450, 874–878.
Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sasser, M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 1502, 101.
Söhngen, N.L. 1906. Ueber bakterien, welche methan als kohlenstoffnahrung und energiequelle gebrauchen. Zentralbl Bakteriol Parasitenk Infektionskr Hyg. Abt. II 15, 513–517.
Stoecker, K., Bendinger, B., Schoning, B., Nielsen, P.H., Nielsen, J.L., Baranyi, C., Toenshoff, E.R., Daims, H., and Wagner, M. 2006. Cohn’s crenothrix is a filamentous methane oxidizer with an unusual methane monooxygenase. Proc. Natl. Acad. Sci. USA 103, 2363–2367.
Takeuchi, M., Kamagata, Y., Oshima, K., Hanada, S., Tamaki, H., Marumo, K., Maeda, H., Nedachi, M., Hattori, M., Iwasaki, W., et al. 2014. Methylocaldum marinum sp. nov., a thermotolerant, methane-oxidizing bacterium isolated from marine sediments, and emended description of the genus Methylocaldum. Int. J. Syst. Evol. Microbiol. 64, 3240–3246.
Vorobev, A.V., Baani, M., Doronina, N.V., Brady, A.L., Liesack, W., Dunfield, P.F., and Dedysh, S.N. 2011. Methyloferula stellata gen. nov., sp. nov., an acidophilic, obligately methanotrophic bacterium that possesses only a soluble methane monooxygenase. Int. J. Syst. Evol. Microbiol. 61, 2456–2463.
Ward, N., Larsen, O., Sakwa, J., Bruseth, L., Khouri, H., Durkin, A.S., Dimitrov, G., Jiang, L., Scanlan, D., Kang, K.H., et al. 2004. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath). PLoS Biol. 2, e303.
Weisburg, W.G., Barns, S.M., Pelletier, D.A., and Lane, D.J. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703.
Whittenbury, R., Krieg, N.R., and Holt, J.G. 1984. Family IV. methylococcaceae, pp. 256–261. In Krieg, N.R. and Holt, J.G. (eds.), Bergey’s Manual of Systematic Bacteriology, The Williams & Wilkins Co., Baltimore, Maryland, USA.
Widdel, F. and Bak, F. 1992. Gram-negative mesophilic sulfatereducing bacteria, pp. 3352–3378. In Balows, A., Trüper, H.G., Dworkin, M., Harder, W., and Schleifer, K.H. (eds.), The Prokaryotes: A handbook on the biology of bacteria: Ecophysiology, isolation, identification, applications. Springer New York, N.Y., USA.
Yoon, S.H., Ha, S.M., Kwon, S., Lim, J., Kim, Y., Seo, H., and Chun, J. 2017. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplemental material for this article may be found at http://www.springerlink.com/content/120956.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Nguyen, NL., Yu, WJ., Yang, HY. et al. A novel methanotroph in the genus Methylomonas that contains a distinct clade of soluble methane monooxygenase. J Microbiol. 55, 775–782 (2017). https://doi.org/10.1007/s12275-017-7317-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-017-7317-3