Abstract
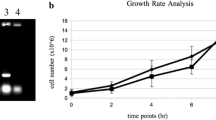
The SRL4 (YPL033C) gene was initially identified by the screening of Saccharomyces cerevisiae genes that play a role in DNA metabolism and/or genome stability using the SOS system of Escherichia coli. In this study, we found that the srl4Δ mutant cells were resistant to the chemicals that inhibit nucleotide metabolism and evidenced higher dNTP levels than were observed in the wild-type cells in the presence of hydroxyurea. The mutant cells also showed a significantly faster growth rate and higher dNTP levels at low temperature (16°C) than were observed in the wild-type cells, whereas we detected no differences in the growth rate at 30°C. Furthermore, srl4Δ was shown to suppress the lethality of mutations of the essential S phase checkpoint genes, RAD53 and LCD1. These results indicate that SRL4 may be involved in the regulation of dNTP production by its function as a negative regulator of ribonucleotide reductase.
Similar content being viewed by others
References
Alvino, G.M., D. Collingwood, J.M. Murphy, J. Delrow, B.J. Brewer, and M.K. Raghuraman. 2007. Replication in hydroxyurea: It’s a matter of time. Mol. Cell Biol. 27, 6396–6406.
Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21, 2239–3330.
Ben-Yehoyada, M., J. Gautier, and A. Dupre. 2007. The DNA damage response during an unperturbed S-phase. DNA Repair 6, 914–922.
Branzei, D. and M. Foiani. 2006. The Rad53 signal transduction pathway: replication fork stabilization, DNA repair, and adaptation. Exp. Cell Res. 312, 2654–2659.
Chabes, A., V. Domkin, and L. Thelander. 1999. Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J. Biol. Chem. 274, 36679–36683.
Chabes, A. and B. Stillman. 2007. Constitutively high dNTP concentration inhibits cell cycle progression and the DNA damage checkpoint in yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 104, 1183–1188.
Chen, S., M.B. Smolka, and H. Zhou. 2007. Mechanism of Dun1 activation by Rad53 phosphorylation in Saccharomyces cerevisiae. J. Biol. Chem. 282, 986–995.
Chris, K., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics, p. 207–217. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, NY, USA.
Fu, Y. and W. Xiao. 2006. Identification and characterization of CRT10 as a novel regulator of Saccharomyces cerevisiae ribonucleotide reductase genes. Nucleic Acids Res. 34, 1876–1883.
Gerik, K.J., X. Li, and A. Pautz, and P.M. Burgers. 1998. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 273, 19747–19755.
Guarino, E., I. Salguero, A. Jimenez-Sanchez, and E.C. Guzman. 2007. Double-strand break generation under deoxyribonucleotide starvation in Escherichia coli. J. Bacteriol. 189, 5782–5786.
Hakansson, P., L. Dahl, O. Chilkova, V. Domkin, and L. Thelander. 2006. The Schizosaccharomyces pombe replication inhibitor Spd1 regulates ribonucleotide reductase activity and dNTPs by binding to the large Cdc22 subunit. J. Biol. Chem. 281, 1778–1783.
Huang, M., Z. Zhou, and S.J. Elledge. 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94, 595–605.
Koc, A. and G.F. Merrill. 2007. Checkpoint deficient rad53-11 yeast cannot accumulate dNTPs in response to DNA damage. Biochem. Biophys. Res. Commun. 353, 527–530.
Longhese, M.P., M. Clerici, and G. Lucchini. 2003. The S-phase checkpoint and its regulation in Saccharomyces cerevisiae. Mutat. Res. 532, 41–58.
Muller, R., D. Mumberg, and F.C. Lucibello. 1993. Signals and genes in the control of cell-cycle progression. Biochim. Biophys. Acta 1155, 151–179.
Nordlund, P. and P. Reichard. 2006. Ribonucleotide reductases. Annu. Rev. Biochem. 75, 681–706.
North, T.W., R.K. Bestwick, and C.K. Mathews. 1980. Detection of activities that interfere with the enzymatic assay of deoxyribonucleoside 5’-triphosphates. J. Biol. Chem. 255, 6640–6645.
Perkins, E., J.F. Sterling, V.I. Hashem, and M.A. Resnick. 1999. Yeast and human genes that affect the Escherichia coli SOS response. Proc. Natl. Acad. Sci. USA 96, 2204–2209.
Rouse, J. and S.P. Jackson. 2000. LCD1: an essential gene involved in checkpoint control and regulation of the MEC1 signaling pathway in Saccharomyces cerevisiae. EMBO J. 19, 5801–5812.
Sikorski, R. and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27.
Thomas, B.J. and R. Rothstein. 1989. The genetic control of directrepeat recombination in Saccharomyces: the effect of rad52 and rad1 on mitotic recombination at GAL10, a transcriptionally regulated gene. Genetics 123, 725–738.
Zhang, Z., K. Yang, C.C. Chen, J. Feser, and M. Huang. 2007. Role of the C terminus of the ribonucleotide reductase large subunit in enzyme regeneration and its inhibition by Sml1. Proc. Natl. Acad. Sci. USA 104, 2217–2222.
Zhao, X., E.G.D. Muller, and R. Rothstein. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2, 329–340.
Zhao, X., A. Chabes, V. Domkin, L. Thelander, and R. Rothstein. 2001. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 20, 3544–3553.
Zhao, X. and R. Rothstein. 2002. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc. Natl. Acad. Sci. USA 99, 3746–3751.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Choi, DH., Oh, YM., Kwon, SH. et al. The mutation of a novel Saccharomyces cerevisiae SRL4 gene rescues the Lethality of rad53 and lcd1 mutations by modulating dNTP levels. J Microbiol. 46, 75–80 (2008). https://doi.org/10.1007/s12275-008-0013-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-008-0013-6