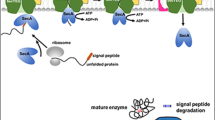
Abstract
Bacillus subtilis has been studied for more than half a century, ever since the dawn of molecular biology, as a representative Gram-positive bacterium and cell factory. Two characteristic capacities of B. subtilis, namely its natural competence for DNA uptake and high-level enzyme secretion, have been investigated and exploited intensively during these long years. As a consequence, this bacterium has evolved into an excellent platform for synthetic biological research and development. In this review, we outline basic concepts for B. subtilis cell factory engineering, and we describe several examples of its applications in the production of proteins and high-value metabolites. In particular, we highlight engineering approaches that can make the already very efficient Bacillus protein secretion pathways even more efficient for the production of enzymes and pharmaceutical proteins. We further showcase examples of metabolic engineering in B. subtilis based on synthetic biology principles to produce various high-value or health-promoting substances, especially inositol stereoisomers. We conclude that the versatile traits of B. subtilis, combined with multi-omics approaches and rapidly developing technologies for genome engineering and high-throughput screening enable us to modify and optimize this bacterium’s metabolic circuits to deliver compounds that are needed for a green and sustainable society as well as a healthy population.
Similar content being viewed by others
References
Perkins, J., M. Wyss, H. P. Hohmann, and U. Sauer (2009) Metabolic engineering of Bacillus subtilis. pp. 908–914. In: C. D. Smolke (ed.). The Metabolic Pathway Engineering Handbook: Fundamentals. CRC press, Boca Raton, FL, USA.
Sonenshein, A. L. (2007) Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5: 917–927.
van Dijl, J. M. and M. Hecker (2013) Bacillus subtilis: from soil bacterium to super-secreting cell factory. Microb. Cell Fact. 12: 3.
Dubnau, D. (1991) Genetic competence in Bacillus subtilis. Microbiol. Rev. 55: 395–424.
Higgins, D. and J. Dworkin (2012) Recent progress in Bacillus subtilis sporulation. FEMS Microbiol. Rev. 36: 131–148.
Westers, H., R. Dorenbos, J. M. van Dijl, J. Kabel, T. Flanagan, K. M. Devine, F. Jude, S. J. Seror, A. C. Beekman, E. Darmon, C. Eschevins, A. de Jong, S. Bron, O. P. Kuipers, A. M. Albertini, H. Antelmann, M. Hecker, N. Zamboni, U. Sauer, C. Bruand, D. S. Ehrlich, J. C. Alonso, M. Salas, and W. J. Quax (2003) Genome engineering reveals large dispensable regions in Bacillus subtilis. Mol. Biol. Evol. 20: 2076–2090.
Caspi, R., T. Altman, R. Billington, K. Dreher, H. Foerster, C. A. Fulcher, T. A. Holland, I. M. Keseler, A. Kothari, A. Kubo, M. Krummenacker, M. Latendresse, L. A. Mueller, Q. Ong, S. Paley, P. Subhraveti, D. S. Weaver, D. Weerasinghe, P. Zhang, and P. D. Karp (2014) The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 42: D459–D471.
Michna, R. H., F. M. Commichau, D. Toedter, C. P. Zschiedrich, and J. Stuelke (2014) SubtiWiki-a database for the model organism Bacillus subtilis that links pathway, interaction and expression information. Nucleic Acids Res. 42: D692–D698.
Sierro, N., Y. Makita, M. de Hoon, and K. Nakai (2008) DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 36: D93–D96.
Pohl, S. and C. R. Harwood (2010) Heterologous protein secretion by Bacillus species from the cradle to the grave. Adv. Appl. Microbiol. 73: 1–25.
Liu, L., Y. Liu, H. D. Shin, R. R. Chen, N. S. Wang, J. Li, G. Du, and J. Chen (2013) Developing Bacillus spp. as a cell factory for production of microbial enzymes and industrially important biochemicals in the context of systems and synthetic biology. Appl. Microbiol. Biotechnol. 97: 6113–6127.
Tjalsma, H., H. Antelmann, J. D. H. Jongbloed, P. G. Braun, E. Darmon, R. Dorenbos, J. Y. F. Dubois, H. Westers, G. Zanen, W. J. Quax, O. P. Kuipers, S. Bron, M. Hecker, and J. M. van Dijl (2004) Proteomics of protein secretion by Bacillus subtilis: Separating the “secrets” of the secretome. Microbiol. Mol. Biol. Rev. 68: 207–233.
Westers, L., H. Westers, and W. J. Quax (2004) Bacillus subtilis as cell factory for pharmaceutical proteins: a biotechnological approach to optimize the host organism. Biochim. Biophys. Acta. 1694: 299–310.
Jensen, C. L., K. Stephenson, S. T. Jorgensen, and C. Harwood (2000) Cell-associated degradation affects the yield of secreted engineered and heterologous proteins in the Bacillus subtilis expression system. Microbiology. 146: 2583–2594.
Harwood, C. R., J. M. Mouillon, S. Pohl, and J. Arnau (2018) Secondary metabolite production and the safety of industrially important members of the Bacillus subtilis group. FEMS Microbiol. Rev. 42: 721–738.
Hohmann, H. P., J. M. van Dijl, L. Krishnappa, and Z. Prágai (2016) Host organisms: Bacillus subtilis. pp. 221–298. In: C. Wittmann and J. C. Liao (eds). Industrial Biotechnology. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany.
Bolhuis, A., C. P. Broekhuizen, A. Sorokin, M. L. Van Roosmalen, G. Venema, S. Bron, W. J. Quax, and J. M. van Dijl (1998) SecDF of Bacillus subtilis, a molecular siamese twin required for the efficient secretion of proteins. J. Biol. Chem. 273: 21217–21224.
Zimmer, J., Y. Nam, and T. A. Rapoport (2008) Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 455: 936–943.
Furukawa, A., K. Yoshikaie, T. Mori, H. Mori, Y. V. Morimoto, Y. Sugano, S. Iwaki, T. Minamino, Y. Sugita, Y. Tanaka, and T. Tsukazaki (2017) Tunnel formation inferred from the I-form structures of the proton-driven protein secretion motor SecDF. Cell Rep. 19: 895–901.
Bunai, K., H. Takamatsu, T. Horinaka, A. Oguro, K. Nakamura and K. Yamane (1996) Bacillus subtilis Ffh, a homologue of mammalian SRP54, can intrinsically bind to the precursors of secretory proteins. Biochem. Biophys. Res. Commun. 227: 762–767.
Nakamura, K., S. Yahagi, T. Yamazaki, and K. Yamane (1999) Bacillus subtilis histone-like protein, HBsu, is an integral component of a SRP-like particle that can bind the Alu domain of small cytoplasmic RNA. J. Biol. Chem. 274: 13569–13576.
Zanen, G., H. Antelmann, R. Meima, J. D. H. Jongbloed, M. Kolkman, M. Hecker, J. M. van Dijl, and W. J. Quax (2006) Proteomic dissection of potential signal recognition particle dependence in protein secretion by Bacillus subtilis. Proteomics. 6: 3636–3648.
Müller, J. P., J. Ozegowski, S. Vettermann, J. Swaving, K. H. Van Wely, and A. J. Driessen (2000) Interaction of Bacillus subtilis CsaA with SecA and precursor proteins. Biochem. J. 348: 367–373.
Moliere, N. and K. Turgay (2009) Chaperone-protease systems in regulation and protein quality control in Bacillus subtilis. Res. Microbiol. 160: 637–644.
Seydlová, G., P. Halada, R. Fišer, O. Toman, A. Ulrych, and J. Svobodová (2012) DnaK and GroEL chaperones are recruited to the Bacillus subtilis membrane after short-term ethanol stress. J. Appl. Microbiol. 112: 765–774.
Kontinen, V. P., P. Saris, and M. Sarvas (1991) A gene (prsA) of Bacillus subtilis involved in a novel, late stage of protein export. Mol. Microbiol. 5: 1273–1283.
Jacobs, M., J. B. Andersen, V. Kontinen, and M. Sarvas (1993) Bacillus subtilis PrsA is required in vivo as an extracytoplasmic chaperone for secretion of active enzymes synthesized either with or without pro-sequences. Mol. Microbiol. 8: 957–966.
Kontinen, V. P. and M. Sarvas (1993) The PrsA lipoprotein is essential for protein secretion in Bacillus subtilis and sets a limit for high-level secretion. Mol. Microbiol. 8: 727–737.
Vitikainen, M., I. Lappalainen, R. Seppala, H. Antelmann, H. Boer, S. Taira, H. Savilahti, M. Hecker, M. Vihinen, M. Sarvas, and V. P. Kontinen (2004) Structure-function analysis of PrsA reveals roles for the parvulin-like and flanking N- and C-terminal domains in protein folding and secretion in Bacillus subtilis. J. Biol. Chem. 279: 19302–19314.
Kouwen, T. R. H. M., A. van der Goot, R. Dorenbos, T. Winter, H. Antelmann, M. C. Plaisier, W. J. Quax, J. M. van Dijl, and J. Y. F. Dubois (2007) Thiol-disulphide oxidoreductase modules in the low-GC Gram-positive bacteria. Mol. Microbiol. 64: 984–999.
Kouwen, T. R. H. M., J. Y. F. Dubois, R. Freudl, W. J. Quax, and J. M. van Dijl (2008) Modulation of thiol-disulfide oxidoreductases for increased production of disulfide-bond-containing proteins in Bacillus subtilis. Appl. Environ. Microbiol. 74: 7536–7545.
Kouwen, T. R. and J. M. van Dijl (2009) Applications of thioldisulfide oxidoreductases for optimized in vivo production of functionally active proteins in Bacillus. Appl. Microbiol. Biotechnol. 85: 45–52.
Tjalsma, H., A. Bolhuis, J. D. H. Jongbloed, S. Bron, and J. M. Van Dijl (2000) Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64: 515–547.
Tjalsma, H., A. Bolhuis, M. L. Van Roosmalen, T. Wiegert, W. Schumann, C. P. Broekhuizen, W. J. Quax, G. Venema, S. Bron, and J. M. van Dijl (1998) Functional analysis of the secretory precursor processing machinery of Bacillus subtilis: identification of a eubacterial homolog of archaeal and eukaryotic signal peptidases. Genes Dev. 12: 2318–2331.
van Roosmalen, M. L., N. Geukens, J. D. H. Jongbloed, H. Tjalsma, J. Y. F. Dubois, S. Bron, J. M. van Dijl, and J. Anne (2004) Type I signal peptidases of Gram-positive bacteria. Biochim. Biophys. Acta. 1694: 279–297.
Dalbey, R. E., P. Wang, and J. M. van Dijl (2012) Membrane proteases in the bacterial protein secretion and quality control pathway. Microbiol. Mol. Biol. Rev. 76: 311–330.
Heinrich, J., T. Lundén, V. P. Kontinen, and T. Wiegert (2008) The Bacillus subtilis ABC transporter EcsAB influences intramembrane proteolysis through RasP. Microbiology. 154: 1989–1997.
Neef, J., C. Bongiorni, V. J. Goosens, B. Schmidt, and J. M. van Dijl (2017) Intramembrane protease RasP boosts protein production in Bacillus. Microb. Cell Fact. 16: 57.
Bolhuis, A., A. Matzen, H. L. Hyyryläinen, V. P. Kontinen, R. Meima, J. Chapuis, G. Venema, S. Bron, R. Freudl, and J. M. van Dijl (1999) Signal peptide peptidase- and ClpP-like proteins of Bacillus subtilis required for efficient translocation and processing of secretory proteins. J. Biol. Chem. 274: 24585–24592.
Neef, J., C. Bongiorni, B. Schmidt, V. J. Goosens, and J. M. van Dijl (2020) Relative contributions of non-essential Sec pathway components and cell envelope-associated proteases to highlevel enzyme secretion by Bacillus subtilis. Microb. Cell Fact. 19: 52.
Hyyrylainen, H. L., A. Bolhuis, E. Darmon, L. Muukkonen, P. Koski, M. Vitikainen, M. Sarvas, Z. Pragai, S. Bron, J. M. van Dijl, and V. P. Kontinen (2001) A novel two-component regulatory system in Bacillus subtilis for the survival of severe secretion stress. Mol. Microbiol. 41: 1159–1172.
Antelmann, H., E. Darmon, D. Noone, J. W. Veening, H. Westers, S. Bron, O. P. Kuipers, K. M. Devine, M. Hecker, and J. M. van Dijl (2003) The extracellular proteome of Bacillus subtilis under secretion stress conditions. Mol. Microbiol. 49: 143–156.
Vitikainen, M., H. L. Hyyrylainen, A. Kivimaki, V. P. Kontinen, and M. Sarvas (2005) Secretion of heterologous proteins in Bacillus subtilis can be improved by engineering cell components affecting post-translocational protein folding and degradation. J. Appl. Microbiol. 99: 363–375.
Lulko, A. T., J. W. Veening, G. Buist, W. K. Smits, E. J. Blom, A. C. Beekman, S. Bron, and O. P. Kuipers (2007) Production and secretion stress caused by overexpression of heterologous alpha-amylase leads to inhibition of sporulation and a prolonged motile phase in Bacillus subtilis. Appl. Environ. Microbiol. 73: 5354–5362.
Noone, D., A. Howell, R. Collery, and K. M. Devine (2001) YkdA and YvtA, HtrA-like serine proteases in Bacillus subtilis, engage in negative autoregulation and reciprocal cross-regulation of ykdA and yvtA gene expression. J. Bacteriol. 183: 654–663.
Darmon, E., D. Noone, A. Masson, S. Bron, O. P. Kuipers, K. M. Devine, and J. M. van Dijl (2002) A novel class of heat and secretion stress-responsive genes is controlled by the autoregulated CssRS two-component system of Bacillus subtilis. J. Bacteriol. 184: 5661–5671.
Margot, P. and D. Karamata (1996) The wprA gene of Bacillus subtilis 168, expressed during exponential growth, encodes a cell-wall-associated protease. Microbiology. 142: 3437–3444.
Stephenson, K. and C. R. Harwood (1998) Influence of a cell-wall-associated protease on production of α-amylase by Bacillus subtilis. Appl. Environ. Microbiol. 64: 2875–2881.
Bolhuis, A., H. Tjalsma, K. Stephenson, C. R. Harwood, G. Venema, S. Bron, and J. M. van Dijl (1999) Different mechanisms for thermal inactivation of Bacillus subtilis signal peptidase mutants. J. Biol. Chem. 274: 15865–15868.
Krishnappa, L., A. Dreisbach, A. Otto, V. J. Goosens, R. M. Cranenburgh, C. R. Harwood, D. Becher, and J. M. Van Dijl (2013) Extracytoplasmic proteases determining the cleavage and release of secreted proteins, lipoproteins, and membrane proteins in Bacillus subtilis. J. Proteome Res. 12: 4101–4110.
Aguilar Suarez, R., J. Stulke, and J. M. van Dijl (2019) Less is more: toward a genome-reduced Bacillus cell factory for “difficult proteins”. ACS Synth. Biol. 8: 99–108.
Chambert, R., F. Benyahia, and M. F. Petit-Glatron (1990) Secretion of Bacillus subtilis levansucrase. Fe(III) could act as a cofactor in an efficient coupling of the folding and translocation processes. Biochem. J. 265: 375–382.
Hyyrylainen, H. L., M. Vitikainen, J. Thwaite, H. Wu, M. Sarvas, C. R. Harwood, V. P. Kontinen, and K. Stephenson (2000) D-Alanine substitution of teichoic acids as a modulator of protein folding and stability at the cytoplasmic membrane/cell wall interface of Bacillus subtilis. J. Biol. Chem. 275: 26696–26703.
Sarvas, M., C. R. Harwood, S. Bron, and J. M. van Dijl (2004) Post-translocational folding of secretory proteins in Grampositive bacteria. Biochim. Biophys. Acta. 1694: 311–327.
Goosens, V. J., C. G. Monteferrante, and J. M. van Dijl (2014) The Tat system of Gram-positive bacteria. Biochim. Biophys. Acta. 1843: 1698–1706.
Goosens, V. J. and J. M. van Dijl (2016) Twin-arginine protein translocation. pp. 69–94. In: F. Bagnoli and R. Rappuoli (eds.) Protein and Sugar Export and Assembly in Gram-positive Bacteria. Springer International Publishing AG, Cham, Switzerland.
Frain, K. M., C. Robinson, and J. M. van Dijl (2019) Transport of folded proteins by the Tat System. Protein J. 38: 377–388.
Berks, B. C. (1996) A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 22: 393–404.
Chaddock, A. M., A. Mant, I. Karnauchov, S. Brink, R. G. Herrmann, R. B. Klösgen, and C. Robinson (1995) A new type of signal peptide: central role of a twin-arginine motif in transfer signals for the delta pH-dependent thylakoidal protein translocase. EMBO J. 14: 2715–2722.
Jongbloed, J. D. H., U. Martin, H. Antelmann, M. Hecker, H. Tjalsma, G. Venema, S. Bron, J. M. van Dijl, and J. Müller (2000) TatC is a specificity determinant for protein secretion via the twin-arginine translocation pathway. J. Biol. Chem. 275: 41350–41357.
Pop, O., U. Martin, C. Abel, and J. P. Müller (2002) The twin-arginine signal peptide of PhoD and the TatAd/Cd proteins of Bacillus subtilis form an autonomous Tat translocation system. J. Biol. Chem. 277: 3268–3273.
Jongbloed, J. D. H., U. Grieger, H. Antelmann, M. Hecker, R. Nijland, S. Bron, and J. M. van Dijl (2004) Two minimal Tat translocases in Bacillus. Mol. Microbiol. 54: 1319–1325.
Nicolas, P., U. Mäder, E. Dervyn, T. Rochat, A. Leduc, N. Pigeonneau, E. Bidnenko, E. Marchadier, M. Hoebeke, S. Aymerich, D. Becher, P. Bisicchia, E. Botella, O. Delumeau, G. Doherty, E. L. Denham, M. J. Fogg, V. Fromion, A. Goelzer, A. Hansen, E. Härtig, C. R. Harwood, G. Homuth, H. Jarmer, M. Jules, E. Klipp, L. Le Chat, F. Lecointe, P. Lewis, W. Liebermeister, A. March, R. A. T. Mars, P. Nannapaneni, D. Noone, S. Pohl, B. Rinn, F. Rügheimer, P. K. Sappa, F. Samson, M. Schaffer, B. Schwikowski, L. Steil, J. Stülke, T. Wiegert, K. M. Devine, A. J. Wilkinson, J. M. van Dijl, M. Hecker, U. Völker, P. Bessières, and P. Noirot (2012) Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science. 335: 1103–1106.
Goosens, V. J., A. Otto, C. Glasner, C. C. Monteferrante, R. van der Ploeg, M. Hecker, D. Becher, and J. M. van Dijl (2013) Novel twin-arginine translocation pathway-dependent phenotypes of Bacillus subtilis unveiled by quantitative proteomics. J. Proteome Res. 12: 796–807.
Goosens, V. J., C. G. Monteferrante, and J. M. van Dijl (2014) Co-factor insertion and disulfide bond requirements for twin-arginine translocase-dependent export of the Bacillus subtilis Rieske protein QcrA. J. Biol. Chem. 289: 13124–13131.
Monteferrante, C. G., M. Miethke, R. van der Ploeg, C. Glasner, and J. M. van Dijl (2012) Specific targeting of the metallophosphoesterase YkuE to the Bacillus cell wall requires the twin-arginine translocation system. J. Biol. Chem. 287: 29789–29800.
Miethke, M., C. G. Monteferrante, M. A. Marahiel, and J. M. van Dijl (2013) The Bacillus subtilis EfeUOB transporter is essential for high-affinity acquisition of ferrous and ferric iron. Biochim. Biophys. Acta. 1833: 2267–2278.
Eijlander, R. T., J. D. H. Jongbloed, and O. P. Kuipers (2008) Relaxed specificity of the Bacillus subtilis TatAdCd translocase in Tat-dependent protein secretion. J. Bacteriol. 191: 196–202.
Goosens, V. J., A. De-San-Eustaquio-Campillo, R. Carballido-López, and J. M. van Dijl (2015) A Tat ménage à trois — The role of Bacillus subtilis TatAc in twin-arginine protein translocation. Biochim. Biophys. Acta — Mol. Cell Res. 1853: 2745–2753.
Blümmel, A. S., L. A. Haag, E. Eimer, M. Müller, and J. Fröbel (2015) Initial assembly steps of a translocase for folded proteins. Nat. Commun. 6: 7234.
Patel, R., C. Vasilev, D. Beck, C. G. Monteferrante, J. M. van Dijl, C. N. Hunter, C. Smith, and C. Robinson (2014) A mutation leading to super-assembly of twin-arginine translocase (Tat) protein complexes. Biochim. Biophys. Acta — Mol. Cell Res. 1843: 1978–1986.
Kim, J. Y., E. A. Fogarty, F. J. Lu, H. Zhu, G. D. Wheelock, L. A. Henderson, and M. P. DeLisa (2005) Twin-arginine translocation of active human tissue plasminogen activator in Escherichia coli. Appl. Environ. Microbiol. 71: 8451–8459.
Fisher, A. C., J. Y. Kim, R. Perez-Rodriguez, D. Tullman-Ercek, W. R. Fish, L. A. Henderson, and M. P. DeLisa (2008) Exploration of twin-arginine translocation for expression and purification of correctly folded proteins in Escherichia coli. Microb. Biotechnol. 1: 403–415.
Browning, D. F., K. L. Richards, A. R. Peswani, J. Roobol, S. J. W. Busby, and C. Robinson (2017) Escherichia coli “TatExpress” strains super-secrete human growth hormone into the bacterial periplasm by the Tat pathway. Biotechnol. Bioeng. 114: 2828–2836.
Guerrero Montero, I., K. L. Richards, C. Jawara, D. F. Browning, A. R. Peswani, M. Labrit, M. Allen, C. Aubry, E. Davé, D. P. Humphreys, S. J. W. Busby, and C. Robinson (2019) Escherichia coli “TatExpress” strains export several g/L human growth hormone to the periplasm by the Tat pathway. Biotechnol. Bioeng. 116: 3282–3291.
Jongbloed, J. D. H., H. Antelmann, M. Hecker, R. Nijland, S. Bron, U. Airaksinen, F. Pries, W. J. Quax, J. M. van Dijl, and P. G. Braun (2002) Selective contribution of the twin-arginine translocation pathway to protein secretion in Bacillus subtilis. J. Biol. Chem. 277: 44068–44078.
Kolkman, M. A. B., R. van der Ploeg, M. Bertels, M. van Dijk, J. van der Laan, J. M. van Dijl, and E. Ferrari (2008) The twin-arginine signal peptide of Bacillus subtilis YwbN can direct either Tat- or Sec-dependent secretion of different cargo proteins: secretion of active subtilisin via the B. subtilis Tat pathway. Appl. Environ. Microbiol. 74: 7507–7513.
Kouwen, T. R. H. M., R. van der Ploeg, H. Antelmann, M. Hecker, G. Homuth, U. Mäder, and J. M. van Dijl (2009) Overflow of a hyper-produced secretory protein from the Bacillus Sec pathway into the Tat pathway for protein secretion as revealed by proteogenomics. Proteomics. 9: 1018–1032.
van der Ploeg, R., C. G. Monteferrante, S. Piersma, J. P. Barnett, T. R. H. M. Kouwen, C. Robinson, and J. M. van Dijl (2012) High-salinity growth conditions promote Tat-independent secretion of Tat substrates in Bacillus subtilis. Appl. Environ. Microbiol. 78: 7733–7744.
Bolhuis, A., H. Tjalsma, H. E. Smith, A. de Jong, R. Meima, G. Venema, S. Bron, and J. M. van Dijl (1999) Evaluation of bottlenecks in the late stages of protein secretion in Bacillus subtilis. Appl. Environ. Microbiol. 65: 2934–2941.
Collier, D. N. (1994) Expression of Escherichia coli SecB in Bacillus subtilis facilitates secretion of the SecB-dependent maltose-binding protein of E. coli. J. Bacteriol. 176: 4937–4940.
Diao, L., Q. Dong, Z. Xu, S. Yang, J. Zhou, and R. Freudl (2012) Functional implementation of the posttranslational SecB-SecA protein-targeting pathway in Bacillus subtilis. Appl. Environ. Microbiol. 78: 651–659.
Kakeshita, H., Y. Kageyama, K. Ara, K. Ozaki, and K. Nakamura (2010) Enhanced extracellular production of heterologous proteins in Bacillus subtilis by deleting the C-terminal region of the SecA secretory machinery. Mol. Biotechnol. 46: 250–257.
Van Wely, K. H. M., J. Swaving, C. P. Broekhuizen, M. Rose, W. J. Quax, and A. J. M. Driessen (1999) Functional identification of the product of the Bacillus subtilis yvaL gene as a SecG homologue. J. Bacteriol. 181: 1786–1792.
Chen, J., G. Fu, Y. Gai, P. Zheng, D. Zhang, and J. Wen (2015) Combinatorial Sec pathway analysis for improved heterologous protein secretion in Bacillus subtilis: identification of bottlenecks by systematic gene overexpression. Microb. Cell. Fact. 14: 92.
van Dijl, J. M., A. de Jong, J. Vehmaanpera, G. Venema, and S. Bron (1992) Signal peptidase I of Bacillus subtilis: patterns of conserved amino acids in prokaryotic and eukaryotic type I signal peptidases. EMBO J. 11: 2819–2828.
Bolhuis, A., A. Sorokin, V. Azevedo, S. D. Ehrlich, P. G. Braun, A. De Jong, G. Venema, S. Bron, and J. M. van Dijl (1996) Bacillus subtilis can modulate its capacity and specificity for protein secretion through temporally controlled expression of the sipS gene for signal peptidase I. Mol. Microbiol. 22: 605–618.
Meijer, W. J., A. de Jong, G. Bea, A. Wisman, H. Tjalsma, G. Venema, S. Bron, and J. M. van Dijl (1995) The endogenous Bacillus subtilis (natto) plasmids pTA1015 and pTA1040 contain signal peptidase-encoding genes: identification of a new structural module on cryptic plasmids. Mol. Microbiol. 17: 621–631.
Tjalsma, H., M. A. Noback, S. Bron, G. Venema, K. Yamane, and J. M. van Dijl (1997) Bacillus subtilis contains four closely related type I signal peptidases with overlapping substrate specificities. J. Biol. Chem. 272: 25983–25992.
Bron, S., A. Bolhuis, H. Tjalsma, S. Holsappel, G. Venema, and J. M. van Dijl (1998) Protein secretion and possible roles for multiple signal peptidases for precursor processing in Bacilli. J. Biotechnol. 64: 3–13.
Chen, J., Y. Gai, G. Fu, W. Zhou, D. Zhang, and J. Wen (2015) Enhanced extracellular production of α-amylase in Bacillus subtilis by optimization of regulatory elements and overexpression of PrsA lipoprotein. Biotechnol. Lett. 37: 899–906.
Ma, R. J., Y. H. Wang, L. Liu, L. L. Bai, and R. Ban (2018) Production enhancement of the extracellular lipase LipA in Bacillus subtilis: Effects of expression system and Sec pathway components. Protein Expr. Purif. 142: 81–87.
Yang, T., K. Irene, H. Liu, S. Liu, X. Zhang, M. Xu, and Z. Rao (2019) Enhanced extracellular gamma glutamyl transpeptidase production by overexpressing of PrsA lipoproteins and improving its mRNA stability in Bacillus subtilis and application in biosynthesis of L-theanine. J. Biotechnol. 302: 85–91.
Zhang, C., T. Tao, Q. Ying, D. Zhang, F. Lu, X. Bie, and Z. Lu (2012) Extracellular production of lipoxygenase from Anabaena sp. PCC 7120 in Bacillus subtilis and its effect on wheat protein. Appl. Microbiol. Biotechnol. 94: 949–958.
Wu, X. C., S. C. Ng, R. I. Near, and S. L. Wong (1993) Efficient production of a functional single-chain antidigoxin antibody via an engineered Bacillus subtilis expression-secretion system. Biotechnology. 11: 71–76.
Wu, S. C., R. Ye, X. C. Wu, S. C. Ng, and S. L. Wong (1998) Enhanced secretory production of a single-chain antibody fragment from Bacillus subtilis by coproduction of molecular chaperones. J. Bacteriol. 180: 2830–2835.
Wu, S. C., J. C. Yeung, Y. Duan, R. Ye, S. J. Szarka, H. R. Habibi, and S. L. Wong (2002) Functional production and characterization of a fibrin-specific single-chain antibody fragment from Bacillus subtilis: effects of molecular chaperones and a wall-bound protease on antibody fragment production. Appl. Environ. Microbiol. 68: 3261–3269.
Kakeshita, H., Y. Kageyama, K. Endo, M. Tohata, K. Ara, K. Ozaki, and K. Nakamura (2011) Secretion of biologically-active human interferon-β by Bacillus subtilis. Biotechnol. Lett. 33: 1847–1852.
Williams, R. C., M. L. Rees, M. F. Jacobs, Z. Pragai, J. E. Thwaite, L. W. J. Baillie, P. T. Emmerson, and C. R. Harwood (2003) Production of Bacillus anthracis protective antigen is dependent on the extracellular chaperone, PrsA. J. Biol. Chem. 278: 18056–18062.
Quesada-Ganuza, A., M. Antelo-Varela, J. C. Mouritzen, J. Bartel, D. Becher, M. Gjermansen, P. F. Hallin, K. F Appel, M. Kilstrup, M. D. Rasmussen, and A. K. Nielsen (2019) Identification and optimization of PrsA in Bacillus subtilis for improved yield of amylase. Microb. Cell Fact. 18: 158.
Kouwen, T. R. H. M. and J. M. van Dijl (2009) Interchangeable modules in bacterial thiol-disulfide exchange pathways. Trends Microbiol. 17: 6–12.
Dorenbos, R., T. Stein, J. Kabel, C. Bruand, A. Bolhuis, S. Bron, W. J. Quax, and J. M. van Dijl (2002) Thiol-disulfide oxidoreductases are essential for the production of the lantibiotic sublancin 168. J. Biol. Chem. 277: 16682–16688.
Meima, R., C. Eschevins, S. Fillinger, A. Bolhuis, L. W. Hamoen, R. Dorenbos, W. J. Quax, J. M. van Dijl, R. Provvedi, I. Chen, D. Dubnau, and S. Bron (2002) The bdbDC operon of Bacillus subtilis encodes thiol-disulfide oxidoreductases required for competence development. J. Biol. Chem. 277: 6994–7001.
Draskovic, I. and D. Dubnau (2005) Biogenesis of a putative channel protein, ComEC, required for DNA uptake: membrane topology, oligomerization and formation of disulphide bonds. Mol. Microbiol. 55: 881–896.
Bolhuis, A., G. Venema, W. J. Quax, S. Bron, and J. M. van Dijl (1999) Functional analysis of paralogous thiol-disulfide oxidoreductases in Bacillus subtilis. J. Biol. Chem. 274: 24531–24538.
Erlendsson, L. S. and L. Hederstedt (2002) Mutations in the thiol-disulfide oxidoreductases BdbC and BdbD can suppress cytochrome c deficiency of CcdA-defective Bacillus subtilis cells. J. Bacteriol. 184: 1423–1429.
Erlendsson, L. S., R. M. Acheson, L. Hederstedt, and N. E. Le Brun (2003) Bacillus subtilis ResA is a thiol-disulfide oxidoreductase involved in cytochrome c synthesis. J. Biol. Chem. 278: 17852–17858.
Kawamura, F. and R. H. Doi (1984) Construction of a Bacillus subtilis double mutant deficient in extracellular alkaline and neutral proteases. J. Bacteriol. 160: 442–444.
Westers, L., D. S. Dijkstra, H. Westers, J. M. van Dijl, and W. J. Quax (2006) Secretion of functional human interleukin-3 from Bacillus subtilis. J. Biotechnol. 123: 211–224.
Westers, L., H. Westers, G. Zanen, H. Antelmann, M. Hecker, D. Noone, K. M. Devine, J. M. van Dijl, and W. J. Quax (2008) Genetic or chemical protease inhibition causes significant changes in the Bacillus subtilis exoproteome. Proteomics. 8: 2704–2713.
Luo, Z., Q. Gao, X. Li, and J. Bao (2014) Cloning of LicB from Clostridium thermocellum and its efficient secretive expression of thermostable beta-1,3-1,4-glucanase. Appl. Biochem. Biotechnol. 173: 562–570.
Pohl, S., G. Bhavsar, J. Hulme, A. E. Bloor, G. Misirli, M. W. Leckenby, D. S. Radford, W. Smith, A. Wipat, E. D. Williamson, C. R. Harwood, and R. M. Cranenburgh (2013) Proteomic analysis of Bacillus subtilis strains engineered for improved production of heterologous proteins. Proteomics. 13: 3298–3308.
Krishnappa, L., C. G. Monteferrante, J. Neef, A. Dreisbach, and J. M. van Dijl (2014) Degradation of extracytoplasmic catalysts for protein folding in Bacillus subtilis. Appl. Environ. Microbiol. 80: 1463–1468.
Yamamoto, H., S. Kurosawa, and J. Sekiguchi (2003) Localization of the vegetative cell wall hydrolases LytC, LytE, and LytF on the Bacillus subtilis cell surface and stability of these enzymes to cell wall-bound or extracellular proteases. J. Bacteriol. 185: 6666–6677.
Ara, K., K. Ozaki, K. Nakamura, K. Yamane, J. Sekiguchi, and N. Ogasawara (2007) Bacillus minimum genome factory: effective utilization of microbial genome information. Biotechnol. Appl. Biochem. 46: 169–178.
Manabe, K., Y. Kageyama, T. Morimoto, T. Ozawa, K. Sawada, K. Endo, M. Tohata, K. Ara, K. Ozaki, and N. Ogasawara (2011) Combined effect of improved cell yield and increased specific productivity enhances recombinant enzyme production in genome-reduced Bacillus subtilis strain MGB874. Appl. Environ. Microbiol. 77: 8370–8381.
Manabe, K., Y. Kageyama, M. Tohata, K. Ara, K. Ozaki, and N. Ogasawara (2012) High external pH enables more efficient secretion of alkaline α-amylase AmyK38 by Bacillus subtilis. Microb. Cell Fact. 11: 74.
Manabe, K., Y. Kageyama, T. Morimoto, E. Shimizu, H. Takahashi, S. Kanaya, K. Ara, K. Ozaki, and N. Ogasawara (2013) Improved production of secreted heterologous enzyme in Bacillus subtilis strain MGB874 via modification of glutamate metabolism and growth conditions. Microb. Cell Fact. 12: 18.
Antelo-Varela, M., R. Aguilar Suárez, J. Bartel, M. Bernal-Cabas, T. Stobernack, T. Sura, J. M. van Dijl, S. Maaß, and D. Becher (2020) Membrane modulation of super-secreting “midiBacillus” expressing the major Staphylococcus aureus antigen — a mass-spectrometry-based absolute quantification approach. Front. Bioeng. Biotechnol. 8: 143.
Li, Y., X. Zhu, X. Zhang, J. Fu, Z. Wang, T. Chen, and X. Zhao (2016) Characterization of genome-reduced Bacillus subtilis strains and their application for the production of guanosine and thymidine. Microb. Cell Fact. 15: 94.
Reuß, D. R., J. Altenbuchner, U. Mäder, H. Rath, T. Ischebeck, P. K. Sappa, A. Thurmer, C. Guerin, P. Nicolas, L. Steil, B. Zhu, I. Feussner, S. Klumpp, R. Daniel, F. M. Commichau, U. Völker, and J. Stülke (2017) Large-scale reduction of the Bacillus subtilis genome: consequences for the transcriptional network, resource allocation, and metabolism. Genome Res. 27: 289–299.
Averesch, N. J. H. and L. J. Rothschild (2019) Metabolic engineering of Bacillus subtilis for production of para-aminobenzoic acid — unexpected importance of carbon source is an advantage for space application. Microb. Biotechnol. 12: 703–714.
Fu, J., G. Huo, L. Feng, Y. Mao, Z. Wang, H. Ma, T. Chen, and X. Zhao (2016) Metabolic engineering of Bacillus subtilis for chiral pure meso-2,3-butanediol production. Biotechnol. Biofuels. 9: 90.
Romero, S., E. Merino, F. Bolívar, G. Gosset, and A. Martinez (2007) Metabolic engineering of Bacillus subtilis for ethanol production: lactate dehydrogenase plays a key role in fermentative metabolism. Appl. Environ. Microbiol. 73: 5190–5198.
Xu, H., C. Teng, and M. Yu (2006) Improvements of thermal property and crystallization behavior of PLLA based multiblock copolymer by forming stereocomplex with PDLA oligomer. Polymer. 47: 3922–3928.
Awasthi, D., L. Wang, M. S. Rhee, Q. Wang, D. Chauliac, L. O. Ingram, and K. T. Shanmugam (2018) Metabolic engineering of Bacillus subtilis for production of D-lactic acid. Biotechnol. Bioeng. 115: 453–463.
Yang, S., Y. Cao, L. Sun, C. Li, X. Lin, Z. Cai, G. Zhang, and H. Song (2019) Modular pathway engineering of Bacillus subtilis to promote de novo biosynthesis of menaquinone-7. ACS Synth. Biol. 8: 70–81.
Revuelta, J. L., R. Ledesma-Amaro, P. Lozano-Martinez, D. Díaz-Fernández, R. M. Buey, and A. Jiménez (2017) Bioproduction of riboflavin: a bright yellow history. J. Ind. Microbiol. Biotechnol. 44: 659–665.
Abdallah, I. I., H. Pramastya, R. van Merkerk, Sukrasno, and W. J. Quax (2019) Metabolic engineering of Bacillus subtilis toward taxadiene biosynthesis as the first committed step for taxol production. Front. Microbiol. 10: 218.
Berridge, M. J. (2009) Inositol trisphosphate and calcium signalling mechanisms. Biochim. Biophys. Acta. 1793: 933–940.
Clements, R. S. and B. Darnell (1980) myo-Inositol content of common foods: development of a high-myo-inositol diet. Am. J. Clin. Nutr. 33: 1954–1967.
Reynolds, J. E. F. (1993) Martindale: The Extra Pharmacopoeia. 30th ed., p. 1379. Pharmaceutical Press, London, UK.
McLaurin, J., M. E. Kierstead, M. E. Brown, C. A. Hawkes, M. H. Lambermon, A. L. Phinney, A. A. Darabie, J. E. Cousins, J. E. French, M. F. Lan, F. Chen, S. S. N. Wong, H. T. J. Mount, P. E. Fraser, D. Westaway, and P. St George-Hyslop (2006) Cyclohexanehexol inhibitors of Abeta aggregation prevent and reverse Alzheimer phenotype in a mouse model. Nat. Med. 12: 801–808.
Larner, J., L. C. Huang, C. F. Schwartz, A. S. Oswald, T. Y. Shen, M. Kinter, G. Z. Tang, and K. Zeller (1988) Rat liver insulin mediator which stimulates pyruvate dehydrogenase phosphatase contains galactosamine and d-chiroinositol. Biochem. Biophys. Res. Commun. 151: 1416–1426.
Iuorno, M. J., D. J. Jakubowicz, J. P. Baillargeon, P. Dillon, R. D. Gunn, G. Allan, and J. E. Nestler (2002) Effects of d-chiro-inositol in lean women with the polycystic ovary syndrome. Endocr. Pract. 8: 417–423.
Yamaoka, M., S. Osawa, T. Morinaga, S. Takenaka, and K. Yoshida (2011) A cell factory of Bacillus subtilis engineered for the simple bioconversion of myo-inositol to scyllo-inositol, a potential therapeutic agent for Alzheimer’s disease. Microb. Cell Fact. 10: 69.
Yoshida, K., M. Yamaguchi, T. Morinaga, M. Kinehara, M. Ikeuchi, H. Ashida, and Y. Fujita (2008) myo-Inositol catabolism in Bacillus subtilis. J. Biol. Chem. 283: 10415–10424.
Morinaga, T., T. Matsuse, H. Ashida, and K. Yoshida (2010) Differential substrate specificity of two inositol transporters of Bacillus subtilis. Biosci. Biotechnol. Biochem. 74: 1312–1314.
Ramaley, R., Y. Fujita, and E. Freese (1979) Purification and properties of Bacillus subtilis inositol dehydrogenase. J. Biol. Chem. 254: 7684–7690.
Yoshida, K., M. Yamaguchi, T. Morinaga, M. Ikeuchi, M. Kinehara, and H. Ashida (2006) Genetic modification of Bacillus subtilis for production of D-chiro-inositol, an investigational drug candidate for treatment of type 2 diabetes and polycystic ovary syndrome. Appl. Environ. Microbiol. 72: 1310–1315.
Yoshida, K., D. Aoyama, I. Ishio, T. Shibayama, and Y. Fujita (1997) Organization and transcription of the myo-inositol operon, iol, of Bacillus subtilis. J. Bacteriol. 179: 4591–4598.
Yoshida, K., Y. Yamamoto, K. Omae, M. Yamamoto, and Y. Fujita (2002) Identification of two myo-inositol transporter genes of Bacillus subtilis. J. Bacteriol. 184: 983–991.
Yoshida, K., T. Shibayama, D. Aoyama, and Y. Fujita (1999) Interaction of a repressor and its binding sites for regulation of the Bacillus subtilis iol divergon. J. Mol. Biol. 285: 917–929.
Kang, D. M., K. Tanaka, S. Takenaka, S. Ishikawa, and K. Yoshida (2017) Bacillus subtilis iolU encodes an additional NADP+-dependent scyllo-inositol dehydrogenase. Biosci. Biotechnol. Biochem. 81: 1026–1032.
Tanaka, K., S. Tajima, S. Takenaka, and K. Yoshida (2013) An improved Bacillus subtilis cell factory for producing scyllo-inositol, a promising therapeutic agent for Alzheimer’s disease. Microb. Cell Fact. 12: 124.
Tanaka, K., A. Natsume, S. Ishikawa, S. Takenaka, and K. Yoshida (2017) A new-generation of Bacillus subtilis cell factory for further elevated scyllo-inositol production. Microb. Cell Fact. 16: 67.
Fujisawa, T., S. Fujinaga, and H. Atomi (2017) An in vitro enzyme system for the production of myo-inositol from starch. Appl. Environ. Microbiol. 83: e00550–17.
Terakawa, A., A. Natsume, A. Okada, S. Nishihata, J. Kuse, K. Tanaka, S. Takenaka, S. Ishikawa, and K. Yoshida (2016) Bacillus subtilis 5′-nucleotidases with various functions and substrate specificities. BMC Microbiol. 16: 249.
Michon, C., C. M. Kang, S. Karpenko, K. Tanaka, S. Ishikawa, and K. Yoshida (2020) A bacterial cell factory converting glucose into scyllo-inositol, a therapeutic agent for Alzheimer’s disease. Commun. Biol. 3: 93.
Buescher, J. M., W. Liebermeister, M. Jules, M. Uhr, J. Muntel, E. Botella, B. Hessling, R. J. Kleijn, L. Le Chat, F. Lecointe, U. Mäder, P. Nicolas, S. Piersma, F. Rügheimer, D. Becher, P. Bessieres, E. Bidnenko, E. L. Denham, E. Dervyn, K. M. Devine, G. Doherty, S. Drulhe, L. Felicori, M. J. Fogg, A. Goelzer, A. Hansen, C. R. Harwood, M. Hecker, S. Hubner, C. Hultschig, H. Jarmer, E. Klipp, A. Leduc, P. Lewis, F. Molina, P. Noirot, S. Peres, N. Pigeonneau, S. Pohl, S. Rasmussen, B. Rinn, M. Schaffer, J. Schnidder, B. Schwikowski, J. M. van Dijl, P. Veiga, S. Walsh, A. J. Wilkinson, J. Stelling, S. Aymerich, and U. Sauer (2012) Global network reorganization during dynamic adaptations of Bacillus subtilis metabolism. Science. 335: 1099–1103.
Goelzer, A. and V. Fromion (2017) Resource allocation in living organisms. Biochem. Soc. Trans. 45: 945–952.
Bulović, A., S. Fischer, M. Dinh, F. Golib, W. Liebermeister, C. Poirier, L. Tournier, E. Klipp, V. Fromion, and A. Goelzer (2019) Automated generation of bacterial resource allocation models. Metab. Eng. 55: 12–22.
Dessalles, R., V. Fromion, and P. Robert (2020) Models of protein production along the cell cycle: An investigation of possible sources of noise. PLoS One. 15: e0226016.
Meyer, A., R. Pellaux, S. Potot, K. Becker, H. P. Hohmann, S. Panke, and M. Held (2015) Optimization of a whole-cell biocatalyst by employing genetically encoded product sensors inside nanolitre reactors. Nat. Chem. 7: 673–678.
Acknowledgement
We thank our coworkers, past and present, and our friends from the international Bacillus community for the many wonderful discussions and collaborations we have enjoyed over many years. Further, JMvD thanks the Engineering Biology Research centre, University of Kobe, for the generous invitation to spend one month in Kobe as a visiting scholar to write this review in collaboration with KY.
The authors declare no conflict of interest.
Neither ethical approval nor informed consent was required for this study.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yoshida, Ki., van Dijl, J.M. Engineering Bacillus subtilis Cells as Factories: Enzyme Secretion and Value-added Chemical Production. Biotechnol Bioproc E 25, 872–885 (2020). https://doi.org/10.1007/s12257-020-0104-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-020-0104-8