Abstract
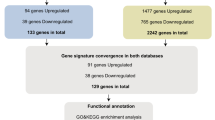
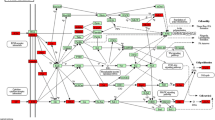
Squamous lung cancer (SQLC) is a common type of lung cancer, but its oncogenesis mechanism is not so clear. The aim of this study was to screen the potential pathways changed in SQLC and elucidate the mechanism of it. Published microarray data of GSE3268 series was downloaded from Gene Expression Omnibus (GEO). Significance analysis of microarrays was performed using software R, and differentially expressed genes (DEGs) were harvested. The functions and pathways of DEGs were mapped in Gene Otology and KEGG pathway database, respectively. A total of 2961 genes were filtered as DEGs between normal and SQLC cells. Cell cycle and metabolism were the mainly changed functions of SQLC cells. Meanwhile genes such as MCM, RFC, FEN1, and POLD may induce SQLC through DNA replication pathway, and genes such as PTTG1, CCNB1, CDC6, and PCNA may be involved in SQLC through cell cycle pathway. It is demonstrated that pathway analysis is useful in the identification of target genes in SQLC.





Similar content being viewed by others
References
Tseng C-Y, Huang Y-C, Su S-Y, Huang J-Y, Lai C-H, Lung C-C, Ho C-C, Liaw Y-P (2012) Cell type specificity of female lung cancer associated with sulfur dioxide from air pollutants in Taiwan: an ecological study. BMC Publ Health 12(1):4
Giangreco A, Lu L, Vickers C, Teixeira VH, Groot KR, Butler CR, Ilieva EV, George PJ, Nicholson AG, Sage EK (2012) β–Catenin determines upper airway progenitor cell fate and preinvasive squamous lung cancer progression by modulating epithelial–mesenchymal transition. J Pathol 226(4):575–587
Sequoia Ecosystem and Recreation Preserve Act of 1999 (1999) (trans: Rep. George E. Brown J). 106th Congress edn
Hammerman P (2012) How far away is targeted treatment for squamous cell lung cancer? Oncology Times UK 9(11):21
Wu M, Tu T, Huang Y, Cao Y (2013) Suppression subtractive hybridization identified differentially expressed genes in lung adenocarcinoma: ERGIC3 as a novel lung cancer-related gene. BMC Cancer 13(1):44
Fang X, Netzer M, Baumgartner C, Bai C, Wang X (2012) Genetic network and gene set enrichment analysis to identify biomarkers related to cigarette smoking and lung cancer. Cancer Treat Rev
Wachi S, Yoneda K, Wu R (2005) Interactome-transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics 21(23):4205–4208
Team RC (2008) R: a language and environment for statistical computing. R Found Stat Comput
Davis S, Meltzer PS (2007) GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 23(14):1846–1847
Diboun I, Wernisch L, Orengo C, Koltzenburg M (2006) Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics 7(1):252
Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3(1):3
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B (Methodological):289–300
Hulsegge I, Kommadath A, Smits MA (2009) Globaltest and GOEAST: two different approaches for Gene Ontology analysis. In: BMC proceedings. BioMed Central Ltd, p S10
Zheng Q, Wang X-J (2008) GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res 36(suppl 2):W358–W363
Zhang JD, Wiemann S (2009) KEGGgraph: a graph approach to KEGG PATHWAY in R and bioconductor. Bioinformatics 25(11):1470–1471
Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, Ullrich RT, Menon R, Maier S, Soltermann A (2010) Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med 2(62):62ra93–62ra93
Tye BK (1999) MCM proteins in DNA replication. Annu Rev Biochem 68(1):649–686
Kikuchi J, Kinoshita I, Shimizu Y, Kikuchi E, Takeda K, Aburatani H, Oizumi S, Konishi J, Kaga K, Matsuno Y (2011) Minichromosome maintenance (MCM) protein 4 as a marker for proliferation and its clinical and clinicopathological significance in non-small cell lung cancer. Lung Cancer 72(2):229–237
Kim YR, Song SY, Kim SS, An CH, Lee SH, Yoo NJ (2010) Mutational and expressional analysis of RFC3, a clamp loader in DNA replication, in gastric and colorectal cancers. Hum Pathol 41(10):1431–1437
Balakrishnan L, Bambara RA (2013) Flap Endonuclease 1. Annu Rev Biochem (0)
Yang M, Guo H, Wu C, He Y, Yu D, Zhou L, Wang F, Xu J, Tan W, Wang G (2009) Functional FEN1 polymorphisms are associated with DNA damage levels and lung cancer risk. Hum Mutat 30(9):1320–1328
Allera-Moreau C, Rouquette I, Lepage B, Oumouhou N, Walschaerts M, Leconte E, Schilling V, Gordien K, Brouchet L, Delisle M (2012) DNA replication stress response involving PLK1, CDC6, POLQ, RAD51 and CLASPIN upregulation prognoses the outcome of early/mid-stage non-small cell lung cancer patients. Oncogenesis 1(10):e30
Li H, Yin C, Zhang B, Sun Y, Shi L, Liu N, Liang S, Lu S, Liu Y, Zhang J (2013) PTTG1 promotes migration and invasion of human non-small cell lung cancer cells and is modulated by miR-186. Carcinogenesis
Andriani F, Roz E, Caserini R, Conte D, Pastorino U, Sozzi G, Roz L (2012) Inactivation of both FHIT and p53 cooperate in deregulating proliferation-related pathways in lung cancer. J Thorac Oncol 7(4):631
Daraselia N, Wang Y, Budoff A, Lituev A, Potapova O, Monforte J, Ossovskaya V (2011) Pathway analysis of primary human non-small cell lung cancer (NSCLC). J Clin Oncol (Meeting Abstracts), p 10573
Yang I-P, Tsai H-L, Hou M-F, Chen K-C, Tsai P-C, Huang S-W, Chou W-W, Wang J-Y, Juo S-HH (2012) MicroRNA-93 inhibits tumor growth and early relapse of human colorectal cancer by affecting genes involved in the cell cycle. Carcinogenesis 33(8):1522–1530
Zhao Y, Li X, Sui X, Tang X, Qin H, Ren H (2010) Expression and significance of PCNA and Caspase-3 in the tissue of lung cancer. Chin J Cell Mol Immunol 26(2):154
Groeger AM, Caputi M, Esposito V, Baldi A, Rossiello R, Santini D, Mancini A, Kaiser HE, Baldi F (2000) Expression of p21 in non small cell lung cancer relationship with PCNA. Anticancer Res 20(5A):3301
Li C, Johnson DE (2013) Liberation of functional p53 by proteasome inhibition in human papilloma virus-positive head and neck squamous cell carcinoma cells promotes apoptosis and cell cycle arrest. Cell Cycle 12(6)
Author information
Authors and Affiliations
Corresponding author
Additional information
Liqiang Qian and Xiaojing Zhao should be regarded as co-first authors.
Rights and permissions
About this article
Cite this article
Qian, L., Luo, Q., Zhao, X. et al. Pathways Enrichment Analysis for Differentially Expressed Genes in Squamous Lung Cancer. Pathol. Oncol. Res. 20, 197–202 (2014). https://doi.org/10.1007/s12253-013-9685-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-013-9685-2