Abstract
Trueperella (T.) bernardiae is a well-known bacterial pathogen in infections of humans, rarely in animals. In the present study, five T. bernardiae isolates, isolated from five Peking ducks of four different farms, were identified by phenotypic properties, by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) analysis, and genotypically by sequencing the 16S ribosomal RNA (rRNA) gene, the superoxide dismutase A encoding gene sodA, and the glyceraldehyde-3-phosphate dehydrogenase encoding gene gap. In addition, the T. bernardiae isolates could be identified with a newly developed loop-mediated isothermal amplification (LAMP) assay based on the gyrase encoding housekeeping gene gyrA. All these tests clearly identified the T. bernardiae isolates to the species level. However, the detection of the specific gene gyrA with the newly designed LAMP assay appeared with a high sensitivity and specificity, and could help to identify this bacterial species in human and animal infections in future. The importance of the T. bernardiae isolates for the clinical condition of the ducks and for the problems at farm level remains unclear.
Similar content being viewed by others
Introduction
Trueperella (T.) bernardiae is a gram-positive, non-motile, and facultatively anaerobic coccobacillus which was originally identified as a coryneform group 2 bacterium (Na’Was et al. 1987). Eight years later, this bacterium was recognized as Actinomyces bernardiae (Funke et al. 1995) and then reclassified as part of the genus Arcanobacterium as Arcanobacterium bernardiae (Ramos et al. 1997). According to a proposal of Yassin et al. (2011), Arcanobacterium bernardiae was finally classified to the newly described genus Trueperella as T. bernardiae, together with T. pyogenes, T. bialowiezensis, T. bonasi, and T. abortisuis.
In humans, T. bernardiae was first described as an opportunistic pathogen and later could be identified alone or in combination with other bacteria as causing joint infections (Gilarranz et al. 2016; Gowe et al. 2018), urinary tract infections (Lepargneur et al. 1998), abscesses (Parha et al. 2015; VanGorder et al. 2016; Calatrava et al. 2019; Pan et al. 2019), wound infections (Weitzel et al. 2011; Rattes et al. 2016; Cobo et al. 2017), a diabetic foot infection (Schneider et al. 2015), bacteremia (Otto et al. 2013; Roh et al. 2019), and septic thrombophlebitis (Lawrence et al. 2018).
The first characterization of T. bernardiae of animal origin (3-day-old piglet) was made by Hijazin et al. (2012c). In a second case, Arnafia et al. (2017) described a T. bernardiae strain recovered from a purulent thelitis of a 12-year-old male dog.
These previously isolated T. bernardiae isolates of animal origin were identified and further characterized by matrix-assisted desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) and by sequencing various genomic targets (Hijazin et al. 2012c; Arnafia et al. 2017).
At present, no further data are available concerning the isolation of T. bernardiae from animals or animal infections.
In the present investigation, five T. bernardiae isolates recovered from post mortem samples of Peking ducks (Anas platyrhynchos domesticus) could be identified phenotypically and genotypically, and with a newly described loop-mediated isothermal amplification (LAMP) assay based on the housekeeping gene gyrA.
Material and methods
Bacterial strains
The five T. bernardiae isolates investigated in the present study (T. bernardiae D12-0613-1-4-3, T. bernardiae D13-1622-5-3-2, T. bernardiae D14-1577-4-8-1, T. bernardiae D13-1772-748-1-2, T. bernardiae D14-1481-2029-1-1) were isolated from five Peking ducks (Anas platyrhynchos domesticus) from three different farms in Germany (n = 3) and one in Thailand (n = 2). The Geman samples were collected from the heart, lung, and joint, while the Thailand samples were delivered as swabs after post-mortem examination; unfortunately, the source organs were not defined. All five isolates were collected during post-mortem examination in the period between 2012 and 2014. Further details about the five isolates are given in Table 1. The bacterial cultivation and a preliminary identification of the bacteria were performed at Ripac-Labor GmbH, Potsdam, Germany. The bacterial culturing of the T. bernardiae isolates was carried out on sheep blood agar plates (Oxoid GmbH, Wesel, Germany) for 48 h at 37 °C under a microaerophilic gas atmosphere using a candle jar.
Phenotypic identification
A phenotypic identification was performed using conventional cultural and biochemical assays as previously shown (Hassan et al. 2009; Ülbegi-Mohyla et al. 2009) and with the API-Coryne test system (BioMérieux Deutschland GmbH, Nürtingen, Germany) in accordance with the manufacturer’s instructions. Furthermore, the bacterial isolates were identified by MALDI-TOF MS using a Microflex LT (Bruker Daltonik GmbH, Bremen, Germany) instrument following the manufacturer’s instructions using the direct transfer method. Briefly, one microbial colony was first smeared in duplicate onto spots of the MALDI MSP 96 target plate (MicroScout Target plate; Bruker Daltonik GmbH) with sterile toothpicks. The air-dried bacteria were overlaid with 1µL of an α-cyan 4-hydroxycinnamic acid matrix solution (HCCA, in 50% acetonitrile and 2.5% trifluoroacetic acid in pure water), followed by drying and loading into the mass spectrometer. The analysis of the spectra was carried out by MBT Compass Explorer 4.1 software (Bruker Daltonik GmbH).
DNA extraction
The genomic DNA of the five isolates, type strain T. bernardiae DSM 9152 T, and various other strains of genus Trueperella and genus Arcanobacterium (A.) were extracted using the DNeasy Blood and Tissue kit (Qiagen GmbH, Hilden, Germany), in conformance with the manufacturer’s instructions. The concentration and purity of DNA were measured by means of a NanoDrop spectrophotometer (ND1000; Thermo Fisher Scientific GmbH, Dreieich, Germany).
Sequencing the molecular targets
The five T. bernardiae isolates were also investigated by sequencing the following molecular targets: 16S rRNA gene, superoxide dismutase A encoding gene sodA, and glyceraldehyde-3-phosphate dehydrogenase encoding gene gap. The sequences of the oligonucleotide primers and the PCR conditions were used as previously described for 16S rRNA gene (Hassan et al. 2009), gene sodA (Hijazin et al. 2011, 2012c), and gene gap (Wickhorst et al. 2019). The PCR products were purified and sequenced by Eurofins Genomics Germany GmbH (Ebersberg, Germany). The obtained sequences of the different genes of the T. bernardiae isolates were aligned and further analyzed using the clustal w method of the MegAlign program version 15 (DNASTAR, Inc., Madison, WI, USA) and compared with the nucleotide sequences of the targets 16S rRNA gene, sodA, and gap of type strain T. bernardiae DSM 9152 T, and type strain of T. pyogenes DSM 20630 T obtained from the NCBI GenBank, and for control purposes from A. haemolyticum DSM 20595 T also obtained from the NCBI GenBank.
LAMP assay
Design of oligonucleotide primers for LAMP assay
Oligonucleotide primers for the T. bernardiae-specific LAMP assay were developed using the gyrase subunit A encoding gene gyrA of T. bernardiae (LNIZ01000002).
The LAMP primers (forward outer primer gyrA-F3, backward outer primer gyrA-B3, forward inner primer gyrA-FIP, backward inner primer gyrA-BIP, forward loop primer gyrA-LoopF, and backward loop primer gyrA-LoopB) were designed using the LAMP designer software (PREMIER Biosoft, San Francisco, CA, USA) (Table 2). The oligonucleotide primers were synthesized by Eurofins Genomics.
LAMP reaction and amplification conditions
In accordance with the manufacturer’s instructions, the LAMP assay based on gene gyrA was carried out with the five T. bernardiae isolates, type strain T. bernardiae DSM 9152 T, and with control strains of genus Trueperella and closely related genus Arcanobacterium. A total volume of 25 µL for each reaction included 15 µL GspSSD isothermal master mix (ISO-001) (OptiGene Ltd., Horsham, UK) and 2.5 µL primer mix (ISO-001; OptiGene Ltd.), gyrA- F3 primer, and gyrA-B3 primer with a final concentration equivalent to 0.2 µmol/L, gyrA-FIP primer, and gyrA-BIP primer with final concentration equivalent to 0.8 µmol/L and gyrA-LoopF Primer and gyrA-LoopB Primer with a final concentration equivalent to 0.4 µmol/L. Subsequently, 5 µL DNA was added as a template. The LAMP assay was run at 70 °C for 20 min with a melting curve analysis step (annealing curve 98 to 80 °C ramping at 0.05 °C/s) in a real-time fluorometer GenieII® (OptiGene Ltd.).
Analytical sensitivity and specificity of the LAMP assay
Determination of the analytic sensitivity of the LAMP assay was performed seven times using a serially diluted DNA (10−1–10−6) isolated from type strain T. bernardiae DSM 9152 T in AE buffer (10 mM Tris–Cl, 0.5 mM EDTA; pH 9.0) with the conditions mentioned above. DNA isolation and concentration were performed as stated above. The amount of DNA ranged from 3.0 ng/µL (10−0) to 3.0 fg/µL (10−6) bacterial DNA. The colony-forming unit (cfu/mL) was subsequently estimated.
The specificity of the LAMP assay was determined using the DNA of T. bernardiae DSM 9152 T and closely related species of genus Trueperella and Arcanobacterium. These included T. pyogenes DSM 20630 T, T. pyogenes DSM 20594, T. pyogenes 59/11, T. abortisuis DSM 19515 T, T. bialowiezensis DSM 17162 T, T. bonasi DSM 17163 T, A. hippocoleae DSM 15539 T, A. pluranimalium DSM 13483 T, and A. phocae DSM 10002 T. The LAMP assay was performed with the optimized LAMP protocol with a run-time of 20 min.
Results and discussion
The phenotypic properties of the five T. bernardiae isolates of duck origin investigated in the present study were almost identical to those of type strain T. bernardiae DSM 9152 T, and to previously characterized T. bernardiae strains of pig and dog origin (Hijazin et al. 2012c; Arnafia et al. 2017). All T. bernardiae isolates gave positive reactions for pyrazinamidase, pyrrolidonyl arylamidase, and α-glucosidase, and reacted negatively in nitrate reduction and for alkaline phosphatase, β-glucuronidase, β-galactosidase, and N-acetyl-β-glucosaminidase. Also, all isolates did not hydrolyze esculin, urea, and gelatine. The isolates also fermented D-glucose, except T. bernardiae D14-1481-2029-1-1 and type strain T. bernardiae DSM 9152 T, D-ribose, D-maltose, and glycogen, but not D-xylose, D-mannitol, D-lactose, and D-saccharose. In addition, all isolates showed a negative catalase reaction (Table 3).
With the additionally performed MALDI-TOF MS analysis, all five isolates were identified to the species level as T. bernardiae with log-score values varying between 1.87 and 2.2 (data not shown). MALDI-TOF MS appeared to be a fast, accurate, and less expensive tool for microbial identification of bacteria, viruses, and fungi (Singhal et al. 2015), also including T. bernardiae (Hijazin et al. 2012a) and various other species of genera Trueperella and Arcanobacterium (Hijazin et al. 2012a, b).
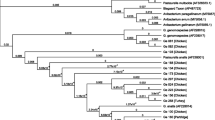
The five T. bernardiae isolates in the current study were additionally identified genotypically by amplification and sequencing of the 16S rRNA gene. The nucleotide sequence of T. bernardiae D12-0613-1-4-3 (GenBank accession number: MT364890), T. bernardiae D13-1622-5-3-2 (MT364891), T. bernardiae D13-1772-748-1-2 (MT364892), T. bernardiae D14-1481-2029-1-1 (MT364893), and T. bernardiae D14-1577-4-8-1 (MT364894) were compared with type strain T. bernardiae DSM 9152 T (HE653979), T. pyogenes DSM 20630 T (X79225), and Arcanobacterium (A.) haemolyticum DSM 20595 T (AJ234059). The nucleotide sequence data of T. bernardiae D12-0613-1-4-3, T. bernardiae D13-1622-5-3-2, T. bernardiae D13-1772-748-1-2, T. bernardiae D14-1481-2029-1-1, and T. bernardiae D14-1577–4-8–1 revealed a sequence homology of 99.7, 99.7, 99.2, 99.0, and 99.7% with type strain T. bernardiae DSM 9152 T, respectively (Fig. 1). The 16S rRNA gene sequence similarities of the five T. bernardiae to T. pyogenes DSM 20630 T and A. haemolyticum DSM 20595 T were equal or less than 98.1 and 94.8%, respectively (Fig. 1).
The five T. bernardiae isolates could be further characterized by PCR-mediated amplification of the genes sodA and gap. The sequences of gene sodA of T. bernardiae D12-0613–1-4–3 (MT410971), T. bernardiae D13-1622–5-3–2 (MT410972), T. bernardiae D13-1772–748-1–2 (MT410973), T. bernardiae D14-1481–2029-1–1 (MT410974), and T. bernardiae D14-1577–4-8–1 (MT410975) resulted in sequence similarities of 95.8, 94.7, 95.5, 93.8, and 95.0% with the sodA gene of type strain T. bernardiae DSM 9152 T (AM989465), respectively, while the similarity within the five isolates was between 99.0 and 100% (Fig. 2a).
Dendrogram analysis of superoxide dismutase A encoding gene sodA (a) and glyceraldehyde-3-phosphate dehydrogenase encoding gene gap (b) of the five T. bernardiae isolates isolated from Peking ducks, type strain T. bernardiae DSM 9152 T, and closely related T. pyogenes DSM 20630 T and A. haemolyticum DSM 20595 T obtained from NCBI GenBank
The additionally investigated gap genes of T. bernardiae D12-0613–1-4–3 (MT410966), T. bernardiae D13-1622–5-3–2 (MT410967), T. bernardiae D13-1772–748-1–2 (MT410968), T. bernardiae D14-1481–2029-1–1 (MT410969), and T. bernardiae D14-1577–4-8–1 (MT410970) showed sequence similarities of 98.5, 98.8, 98.8, 99.0, and 99.0% with the gap gene of type strain T. bernardiae DSM 9152 T (HF947287), respectively, while the similarity within the five isolates was between 99.0 and 99.8% (Fig. 2b). The gene sequences of the sodA and gap genes of the five T. bernardiae isolates showed a clear difference to the control strains T. pyogenes DSM 20630 T (AM949566 and HF947285, respectively) and A. haemolyticum DSM 20595 T (AM983534 and CP002045, respectively) (Fig. 2a, b). All three mentioned genomic targets were already used to characterize various species of genus Trueperella, also including T. bernardiae (Hassan et al. 2009; Hijazin et al. 2011, 2012c; Arnafia et al. 2017).
The additionally used T. bernardiae gyrA-specific LAMP assay could successfully be used to identify the species-specific gene gyrA of all five T. bernardiae isolates in the present investigation. This newly established assay demonstrated a specificity for T. bernardiae DSM 9152 T with an annealing temperature between 91.9 and 92.4 °C. No cross-reactivity with any other related species of genus Trueperella or genus Arcanobacterium could be observed (Table 4).
The developed LAMP assay provided an analytic sensitivity of 30 fg/μL with a mean detection time between 00:08:58 (3.0 ng/μL) and 00:18:35 min (30 fg/μL) (Table 5). For the DNA concentration at 30 fg/μL (10−5), the assay gave positive results in six of seven reactions (85.7%), whereas for DNA concentration at 3.0 fg/µL (10−6), the positive assay resulted in two of seven replicates (28.6%). The results of the T. bernardiae gyrA LAMP assay are shown in Fig. 3 and Table 4.
Positive LAMP assay of the five T. bernardiae isolates T. bernardiae D12-0613–1-4–3, T. bernardiae D13-1622–5-3–2, T. bernardiae D13-1772–748-1–2, T. bernardiae D14-1481–2029-1–1, T. bernardiae D14-1577–4-8–1 obtained from Peking ducks, T. bernardiae DSM 9152 T, and as LAMP negative control T. pyogenes DSM 20630 T and nuclease free water as negative control
The application of LAMP assays as being a rapid and reliable method for detecting species of genus Trueperella and Arcanobacterium has been previously published. In 2013, Zhang et al. developed a LAMP assay using the gene encoding pyolysin, the plo gene, for a specific identification of T. pyogenes (Zhang et al. 2013). Furthermore, a pla LAMP assay was used for identifying A. pluranimalium (Abdulmawjood et al. 2015), and a cpn60 LAMP assay for identifying T. pyogenes from different animal origins (Abdulmawjood et al. 2016; Ahmed et al. 2020; Alssahen et al. 2020).
The present study gives a reliable phenotypic and genotypic characterization of T. bernardiae of duck origin, also including a newly developed LAMP assay. To our knowledge, the study gives the first detailed characterization of this bacterial species isolated from Peking ducks. However, the pathogenic importance of T. bernardiae, which was partly isolated together with various other bacteria from apparently healthy animals, for the high mortality rate or joint infections of the Peking ducks at farm level remains unclear. The described LAMP assay might help to identify this bacterial species in future and might elucidate the role this species plays in human and animal infections.
Availability of data and materials
The data that support the findings of this study are available on NCBI’s Genbank and are accessible through the accession numbers listed in the manuscript.
References
Abdulmawjood A, Wickhorst J, Hashim O, Sammra O, Hassan AA, Alssahen M, Lämmler C, Prenger-Berninghoff E, Klein G (2016) Application of a loop-mediated isothermal amplification (LAMP) assay for molecular identification of Trueperella pyogenes isolated from various origins. Mol Cell Probe 30:205–210
Abdulmawjood A, Wickhorst J, Sammra O, Lämmler C, Foster G, Wragg PN, Prenger-Berninghoff E, Klein G (2015) Development of a loop-mediated isothermal amplification (LAMP) assay for rapid and sensitive identification of Arcanobacterium pluranimalium. Mol Cell Probe 29:468–472
Ahmed MFE, Alssahen M, Lämmler C, Eisenberg T, Plötz M, Abdulmawjood A (2020) Studies on Trueperella pyogenes isolated from an okapi (Okapia johnstoni) and a royal python (Python regius). BMC Vet Res 16:292
Alssahen M, Peters M, Rau J, Hassan AA, Sammra O, Lämmler C, Prenger-Berninghoff E, Plötz M, Abdulmawjood A (2020) Phenotypic and genotypic approach to characterize a Trueperella pyogenes strain isolated from an Eurasian Lynx (Lynx lynx). Berl Münch Tierärzt Wochenschr 133. https://doi.org/10.2376/0005-9366-19037
Arnafia W, Ningrum SG, Sammra O, Alssahen M, Wickhorst J-P, Lämmler C, Prenger-Berninghoff E, Timke M, Abdulmawjood A (2017) Phynotypic and genotypic analysis of a Trueperella bernardiae strain isolated from a dog. Veterinaria (Sarajevo) 66:66–71
Calatrava E, Borrego J, Cobo F (2019) Breast abscess due to Trueperella bernardiae and Actinotignum sanguinis. Rev Esp Quim 32:200–202
Cobo F, Rodriguez-Granger J, Sampedro A, Gutierrez-Fernandez J, Navarro-Mari JM (2017) Two rare cases of wound infections caused by Trueperella bernardiae. Jpn J Infect Dis 70:682–684
Funke G, Ramos CP, Fernandez-Garayzabal JF, Weiss N, Collins MD (1995) Description of human-derived centers for disease control coryneform group 2 bacteria as Actinomyces bernardiae sp. nov. Int J Syst Evol Microbiol 45:57–60
Gilarranz R, Chamizo F, Horcajada I, Bordes-Benitez A (2016) Prosthetic joint infection caused by Trueperella bernardiae. J Infect Chemother 22:642–644
Gowe I, Parsons C, Best M, Parsons E, Prechter S, Vickery S (2018) Successful treatment of olecranon bursitis caused by Trueperella bernardiae: importance of environmental exposure and pathogen identification. Case Rep Infect Dis 2018:5353085
Hassan AA, Ülbegi-Mohyla H, Kanbar T, Alber J, Lämmler C, Abdulmawjood A, Zschöck M, Weiss R (2009) Phenotypic and genotypic characterization of Arcanobacterium haemolyticum isolates from infections of horses. J Clin Microbiol 47:124–128
Hijazin M, Alber J, Lämmler C, Weitzel T, Hassan A, Timke M, Kostrzewa M, Prenger-Berninghoff E, Zschöck M (2012a) Identification of Trueperella (Arcanobacterium) bernardiae by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry analysis and by species-specific PCR. J Med Microbiol 61:457–459
Hijazin M, Hassan AA, Alber J, Lämmler C, Timke M, Kostrzewa M, Prenger-Berninghoff E, Zschöck M (2012b) Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) for species identification of bacteria of genera Arcanobacterium and Trueperella. Vet Microbiol 157:243–245
Hijazin M, Metzner M, Erhard M, Nagib S, Alber J, Lämmler C, Hassan AA, Prenger-Berninghoff E, Zschöck M (2012c) First description of Trueperella (Arcanobacterium) bernardiae of animal origin. Vet Microbiol 159:515–518
Hijazin M, Ülbegi-Mohyla H, Alber J, Lämmler C, Hassan AA, Abdulmawjood A, Prenger-Berninghoff E, Weiss R, Zschöck M (2011) Molecular identification and further characterization of Arcanobacterium pyogenes isolated from bovine mastitis and from various other origins. J Dairy Sci 94:1813–1819
Lawrence C, Waseem S, Newsholme W, Klein J (2018) Trueperella bernardiae: an unusual cause of septic thrombophlebitis in an injection drug user. New Microbes New Infect 26:89–91
Lepargneur J, Heller R, Soulie R, Riegel P (1998) Urinary tract infection due to Arcanobacterium bernardiae in a patient with a urinary tract diversion. Eur J Clin Microbiol Infect Dis 17:399–401
Na’Was T E, Hollis D, Moss C W, Weaver R E, (1987) Comparison of biochemical, morphologic, and chemical characteristics of centers for disease control fermentative coryneform groups 1, 2, and A-4. J Clin Microbiol 25:1354–1358
Otto MP, Foucher B, Lions C, Dardare E, Gerome P (2013) Trueperella bernardiae soft tissue infection and bacteremia. Med Maladies Infect 43:487–489
Pan J, Ho AL, Pendharkar AV, Sussman ES, Casazza M, Cheshier SH, Grant GA (2019) Brain abscess caused by Trueperella bernardiae in a child. Surg Neurol Int 10:35
Parha E, Alalade A, David K, Kaddour H, Degun P, Namnyak S (2015) Brain abscess due to Trueperella bernardiae. Br J Neurosurg 29:728–729
Ramos CP, Foster G, Collins MD (1997) Phylogenetic analysis of the genus Actinomyces based on 16S rRNA gene sequences: description of Arcanobacterium phocae sp. nov., Arcanobacterium bernardiae comb. nov., and Arcanobacterium pyogenes comb. nov. Int J Syst Evol Microbiol 47:46–53
Rattes AL, Araujo MR, Federico MP, Magnoni CD, Neto PA, Furtado GH (2016) Trueperella bernardiae: first report of wound infection post laparoscopic surgery. Clin Case Rep 4:812–815
Roh J, Kim M, Kim D, Yong D, Lee K (2019) First case of Trueperella bernardiae bacteremia in an immunocompromised patient in Korea. Ann Lab Med 39:593–595
Schneider UV, Ekenberg C, Sode N, Knudsen JD (2015) A case of diabetic foot ulcers complicated by severe infection and sepsis with Trueperella bernardiae. JMM Case Reports 2(1). https://doi.org/10.1099/jmmcr.0.000006
Singhal N, Kumar M, Kanaujia PK, Virdi JS (2015) MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol 6:791. https://doi.org/10.3389/fmicb.2015.00791
Ülbegi-Mohyla H, Hassan AA, Kanbar T, Alber J, Lämmler C, Prenger-Berninghoff E, Weiss R, Siebert U, Zschöck M (2009) Synergistic and antagonistic hemolytic activities of bacteria of genus Arcanobacterium and CAMP-like hemolysis of Arcanobacterium phocae and Arcanobacterium haemolyticum with Psychrobacter phenylpyruvicus. Res Vet Sci 87:186–188
VanGorder BRF, Ahmed SS, Rawling RA, Granato PA (2016) Trueperella bernardiae abscess infection: a case report. Clin Microbiol Newsl 12:100–101
Weitzel T, Braun S, Porte L (2011) Arcanobacterium bernardiae bacteremia in a patient with deep soft tissue infection. Surg Infect (Larchmt) 12:83–84
Wickhorst JP, Hassan AA, Sammra O, Alssahen M, Lämmler C, Prenger-Berninghoff E, Naggert M, Timke M, Rau J, Abdulmawjood A (2019) First report on the isolation of Trueperella abortisuis from companion animals. Res Vet Sci 125:465–467
Yassin A, Hupfer H, Siering C, Schumann P (2011) Comparative chemotaxonomic and phylogenetic studies on the genus Arcanobacteriem Collins et al. 1982 emend. Lehnen et al. 2006: proposal for Trueperella gen. nov. and emended description of the genus Arcanobacterium. Int J Syst Evol Microbiol 61:1265–1274
Zhang WL, Meng XL, Wang JW (2013) Sensitive and rapid detection of Trueperella pyogenes using loop-mediated isothermal amplification method. J Microbiol Meth 93:124–126
Funding
Open Access funding enabled and organized by Projekt DEAL. This publication was supported by Deutsche Forschungsgemeinschaft and University of Veterinary Medicine Hannover, Foundation within the funding program Open Access Publishing.
Author information
Authors and Affiliations
Contributions
M.F.E.A, M.A., C.L., A.A., and M.P. contributed to the design of the study, collected, and analyzed the data. B.K., M.M. performed the initial examination of the isolates. M.F.E.A and A.A. drafted the manuscript. C.L. M.M. and M.P. review and editing the manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study did not require official or institutional ethical approval. The material was collected post-mortem and/or during routine diagnosis. According to competent authorities, this kind of research does not require ethics approval or general approval with respect to German law.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ahmed, M.F.E., Alssahen, M., Lämmler, C. et al. Identification of Trueperella bernardiae isolated from peking ducks (Anas platyrhynchos domesticus) by phenotypical and genotypical investigations and by a newly developed loop-mediated isothermal amplification (LAMP) assay. Folia Microbiol 67, 277–284 (2022). https://doi.org/10.1007/s12223-021-00927-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-021-00927-4