Abstract
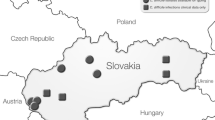
This study aimed to implement a toxigenic culture as an optional third diagnostic step for glutamate dehydrogenase (GDH)-positive and toxin A/B-negative diarrheal stool samples into a diagnostic algorithm for Clostridioides (Clostridium) difficile infection (CDI), and to characterise C. difficile isolates for epidemiological purposes. During the 5-month study, 481 diarrhoeal stool samples from three Slovak hospitals were investigated and 66 non-duplicated GDH-positive stool samples were found. Of them, 36 were also toxin A/B-positive. Twenty-three GDH-positive and toxin A/B-negative stool samples were shown subsequently to be positive following toxigenic culture (TC). Molecular characterisation of C. difficile isolates showed the predominance of PCR ribotype (RT) 001 (n = 37, 56.1%) and the occurrence of RT 176 (n = 3, 4.5%). C. difficile RT 001 isolates clustered to eight clonal complexes (CCs) using multiple-locus variable-number tandem repeats analysis (MLVA). Interestingly, one third of RT 001 isolates clustering in these CCs were cultured from toxin A/B-negative stool samples. Our observations highlight the need of use multiple step diagnostic algorithm in CDI diagnosis in order to detect all CDI cases and to avoid the spread of C. difficile in healthcare settings.


Similar content being viewed by others
References
Bidet P, Barbut F, Lalande V, Burghoffer B, Petit JC (1999) Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol Lett 175:261–266
Bouza E, Peláez T, Alonso R, Catalán P, Muñoz P, Créixems MR (2001) “Second-look” cytotoxicity: an evaluation of culture plus cytotoxin assay of Clostridium difficile isolates in the laboratory diagnosis of CDAD. J Hosp Infect 48(3):233–237
Collins J, Robinson C, Danhof H, Knetsch CW, van Leeuwen HC, Lawley TD, Auchtung JM, Britton RA (2018) Dietary trehalose enhances virulence of epidemic Clostridium difficile. Nature 553:291–294. https://doi.org/10.1038/nature25178
Couturier J, Eckert C, Barbut F (2017) Spatio-temporal variability of the epidemic 027 Clostridium difficile strains in France based on MLVA typing. Anaerobe 48:179–183. https://doi.org/10.1016/j.anaerobe.2017.08.007
Crobach MJ, Planche T, Eckert C, Barbut F, Terveer EM, Dekkers OM et al (2016) European Society of Clinical Microbiology and Infectious Diseases: update of the diagnostic guidance document for Clostridium difficile infection. Clin Microbiol Infect 22(Suppl 4):S63–S81. https://doi.org/10.1016/j.cmi.2016.03.010.
Davies KA, Longshaw CM, Davis GL, Bouza E, Barbut F, Barna Z, Delmée M, Fitzpatrick F, Ivanova K, Kuijper E, Macovei IS, Mentula S, Mastrantonio P, von Müller L, Oleastro M, Petinaki E, Pituch H, Norén T, Nováková E, Nyč O, Rupnik M, Schmid D, Wilcox MH (2014) Underdiagnosis of Clostridium difficile across Europe: the European, multicentre, prospective, biannual, point-prevalence study of Clostridium difficile infection in hospitalised patients with diarrhoea (EUCLID). Lancet Infect Dis 14:1208–1219. https://doi.org/10.1016/S1473-3099(14)70991-0
Davies KA, Ashwin H, Longshaw CM, Burns DA, Davis GL, Wilcox MH, EUCLID study group (2016) Diversity of Clostridium difficile PCR ribotypes in Europe: results from the European, multicentre, prospective, biannual, point-prevalence study of Clostridium difficile infection in hospitalised patients with diarrhoea (EUCLID), 2012 and 2013. Euro Surveill 21(29). https://doi.org/10.2807/1560-7917.ES.2016.21.29.30294
Dingle KE, Didelot X, Quan TP, Eyre DW, Stoesser N, Golubchik T, Harding RM, Wilson DJ, Griffiths D, Vaughan A, Finney JM, Wyllie DH, Oakley SJ, Fawley WN, Freeman J, Morris K, Martin J, Howard P, Gorbach S, Goldstein EJC, Citron DM, Hopkins S, Hope R, Johnson AP, Wilcox MH, Peto TEA, Walker AS, Crook DW, del Ojo Elias C, Crichton C, Kostiou V, Giess A, Davies J (2017) Effects of control interventions on Clostridium difficile infection in England: an observational study. Lancet Infect Dis 17(4):411–421. https://doi.org/10.1016/S1473-3099(16)30514-X
Dresler J, Krutova M, Fucikova A, Klimentova J, Hruzova V, Duracova M, Houdkova K, Salovska B, Matejkova J, Hubalek M, Pajer P, Pisa L, Nyc O (2017) Analysis of proteomes released from in vitro cultured eight Clostridium difficile PCR ribotypes revealed specific expression in PCR ribotypes 027 and 176 confirming their genetic relatedness and clinical importance at the proteomic level. Gut Pathog 9:45. https://doi.org/10.1186/s13099-017-0194-9
Fawley WN, Knetsch CW, MacCannell DR, Harmanus C, Du T, Mulvey MR et al (2015) Development and validation of an internationally-standardized, high-resolution capillary gel-based electrophoresis PCR-ribotyping protocol for Clostridium difficile. PLoS One 10(2):e0118150. https://doi.org/10.1371/journal.pone.0118150
Freeman J, Bauer MP, Baines SD, Corver J, Fawley WN, Goorhuis B, Kuijper EJ, Wilcox MH (2010) The changing epidemiology of Clostridium difficile infections. Clin Microbiol Rev 23:529–549
Gateau C, Couturier J, Coia J, Barbut F (2018) How to: diagnose infection caused by Clostridium difficile. Clin Microbiol Infect 24(5):463–468. https://doi.org/10.1016/j.cmi.2017.12.005
Goorhuis A, Legaria MC, van den Berg RJ, Harmanus C, Klaassen CH, Brazier JS et al (2009) Application of multiple-locus variable-number tandem-repeat analysis to determine clonal spread of toxin A-negative Clostridium difficile in a general hospital in Buenos Aires, Argentina. Clin Microbiol Infect 15:1080–1086. https://doi.org/10.1111/j.1469-0691.2009.02759.x
He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, Connor TR, Harris SR, Fairley D, Bamford KB, D’Arc S, Brazier J, Brown D, Coia JE, Douce G, Gerding D, Kim HJ, Koh TH, Kato H, Senoh M, Louie T, Michell S, Butt E, Peacock SJ, Brown NM, Riley T, Songer G, Wilcox M, Pirmohamed M, Kuijper E, Hawkey P, Wren BW, Dougan G, Parkhill J, Lawley TD (2013) Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet 45:109–113. https://doi.org/10.1038/ng.2478
Indra A, Huhulescu S, Schneeweis M, Hasenberger P, Kernbichler S, Fiedler A, Wewalka G, Allerberger F, Kuijper EJ (2008) Characterization of Clostridium difficile isolates using capillary gel electrophoresis-based PCR ribotyping. J Med Microbiol 57(Pt 11):1377–1382. https://doi.org/10.1099/jmm.0.47714-0.
Knetsch CW, Terveer EM, Lauber C, Gorbalenya AE, Harmanus C, Kuijper EJ, Corver J, van Leeuwen HC (2012) Comparative analysis of an expanded Clostridium difficile reference strain collection reveals genetic diversity and evolution through six lineages. Infect Genet Evol 12:1577–1585. https://doi.org/10.1016/j.meegid.2012.06.003
Krutova M, Matejkova J, Nyc O (2014) C. difficile ribotype 027 or 176? Folia Microbiol (Praha) 59:523–526. https://doi.org/10.1007/s12223-014-0323-5
Krutova M, Matejkova J, Kuijper EJ, Drevinek P, Nyc O, Czech Clostridium difficile study group (2016a) Clostridium difficile PCR ribotypes 001 and 176- the common denominator of C. difficile infection epidemiology in the Czech Republic, 2014. Euro Surveill 21(29). https://doi.org/10.2807/1560-7917.ES.2016.21.29.30296
Krutova M, Nyc O, Matejkova J, Allerberger F, Wilcox MH, Kuijper EJ (2016b) Molecular characterisation of Czech Clostridium difficile isolates collected in 2013-2015. Int J Med Microbiol 306:479–485. https://doi.org/10.1016/j.ijmm.2016.07.003
Krutova M, Nyc O, Matejkova J, Kuijper EJ, Jalava J, Mentula S (2018c) The recognition and characterisation of Finnish Clostridium difficile isolates resembling PCR-ribotype 027. J Microbiol Immunol Infect. https://doi.org/10.1016/j.jmii.2017.02.002
Krutova M, Kinross P, Barbut F, Hajdu A, Wilcox MH, Kuijper EJ, Allerberger F, Delmée M, van Broeck J, Vatcheva-Dobrevska R, Dobreva E, Matica B, Pieridou D, Krůtová M, Nyč O, Olesen B, Märtin P, Mentula S, Barbut F, Arvand M, von Müller L, Papaparaskevas J, Pászti J, Hajdu Á, Gudnason T, Burns K, Spigaglia P, Vulāne K, Debacker M, Scicluna E, Melillo T, Kuijper EJ, Crobach MT, Kacelnik O, Astrup E, Pituch H, Oleastro M, Wiuff C, Coia J, Nováková E, Kolman J, Grilc E, Rupnik M, Bouza E, Reigadas E, Åkerlund T, Tschudin-Sutter S, Wilcox MH, Fairley D, Morris T (2018d) How to: surveillance of Clostridium difficile infections. Clin Microbiol Infect 24(5):469–475. https://doi.org/10.1016/j.cmi.2017.12.008
Lawson PA, Citron DM, Tyrrell KL, Finegold SM (2016) Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O’Toole 1935) Prévot 1938. Anaerobe 40:95–99. https://doi.org/10.1016/j.anaerobe.2016.06.008
Mawer DPC, Eyre DW, Griffiths D, Fawley WN, Martin JSH, Quan TP, Peto TEA, Crook DW, Walker AS, Wilcox MH (2017) Contribution to Clostridium difficile transmission of symptomatic patients with toxigenic strains who are fecal toxin negative. Clin Infect Dis 64:1163–1170. https://doi.org/10.1093/cid/cix079
Nyc O, Krutova M, Liskova A, Matejkova J, Drabek J, Kuijper EJ (2015) The emergence of Clostridium difficile PCR-ribotype 001 in Slovakia. Eur J Clin Microbiol Infect Dis 34:1701–1708. https://doi.org/10.1007/s10096-015-2407-9
Persson S, Torpdahl M, Olsen KE (2008) New multiplex PCR method for the detection of Clostridium difficile toxin A (tcdA) and toxin B (tcdB) and the binary toxin (cdtA/cdtB) genes applied to a Danish strain collection. Clin Microbiol Infect 14:1057–1064. https://doi.org/10.1111/j.1469-0691.2008.02092.x
Pituch H, Obuch-Woszczatyński P, Lachowicz D, Wultańska D, Karpiński P, Młynarczyk G, van Dorp SM, Kuijper EJ, the Polish Clostridium difficile Study Group (2015) Hospital-based Clostridium difficile infection surveillance reveals high proportions of PCR ribotypes 027 and 176 in different areas of Poland, 2011 to 2013. Euro Surveill 20(38). https://doi.org/10.2807/1560-7917.ES.2015.20.38.30025
Polivkova S, Krutova M, Petrlova K, Benes J, Nyc O (2016) Clostridium difficile ribotype 176 - a predictor for high mortality and risk of nosocomial spread? Anaerobe 40:35–40. https://doi.org/10.1016/j.anaerobe.2016.05.002.
Reller ME, Lema CA, Perl TM, Cai M, Ross TL, Speck KA, Carroll KC (2007) Yield of stool culture with isolate toxin testing versus a two-step algorithm including stool toxin testing for detection of toxigenic Clostridium difficile. J Clin Microbiol 45(11):3601–3605
Rupnik M, Tambic Andrasevic A, Trajkovska Dokic E, Matas I, Jovanovic M, Pasic S, Kocuvan A, Janezic S (2016) Distribution of Clostridium difficile PCR ribotypes and high proportion of 027 and 176 in some hospitals in four South Eastern European countries. Anaerobe 42:142–144. https://doi.org/10.1016/j.anaerobe.2016.10.005
Tóth J, Urbán E, Osztie H, Benczik M, Indra A, Nagy E, Allerberger F (2016) Distribution of PCR ribotypes among recent Clostridium difficile isolates collected in two districts of Hungary using capillary gel electrophoresis and review of changes in the circulating ribotypes over time. J Med Microbiol 65:1158–1163. https://doi.org/10.1099/jmm.0.000334
Tschudin-Sutter S, Kuijper EJ, Durovic A, Vehreschild MJGT, Barbut F, Eckert C et al (2018) Guidance document for prevention of Clostridium difficile infection in acute healthcare settings. Clin Microbiol Infect. https://doi.org/10.1016/j.cmi.2018.02.020
Valiente E, Dawson LF, Cairns MD, Stabler RA, Wren BW (2012) Emergence of new PCR ribotypes from the hypervirulent Clostridium difficile 027 lineage. J Med Microbiol 61(Pt 1):49–56. https://doi.org/10.1099/jmm.0.036194-0.
van den Berg RJ, Schaap I, Templeton KE, Klaassen CH, Kuijper EJ (2007) Typing and subtyping of Clostridium difficile isolates by using multiple-locus variable-number tandem-repeat analysis. J Clin Microbiol 45:1024–1028
van Dorp SM, Notermans DW, Alblas J, Gastmeier P, Mentula S, Nagy E, Spigaglia P, Ivanova K, Fitzpatrick F, Barbut F, Morris T, Wilcox MH, Kinross P, Suetens C, Kuijper EJ, for the European Clostridium difficile Infection Surveillance Network (ECDIS-Net) project on behalf of all participants (2016) Survey of diagnostic and typing capacity for Clostridium difficile infection in Europe, 2011 and 2014. Euro Surveill 21(29). https://doi.org/10.2807/1560-7917.ES.2016.21.29.30292
Acknowledgements
We would like to thank all members of the ESCMID study group for Clostridium difficile (ESGCD) for their active contribution to the development and publication of the guidance documents for CDI diagnostics, prevention and treatment.
Funding
The characterisation of C. difficile isolates was supported by MH CZ–DRO, University Hospital Motol, Prague, Czech Republic 00064203.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. For this type of study, formal consent is not required.
Rights and permissions
About this article
Cite this article
Krehelova, M., Nyč, O., Sinajová, E. et al. The predominance and clustering of Clostridioides (Clostridium) difficile PCR ribotype 001 isolates in three hospitals in Eastern Slovakia, 2017. Folia Microbiol 64, 49–54 (2019). https://doi.org/10.1007/s12223-018-0629-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-018-0629-9