Abstract
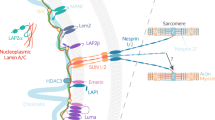
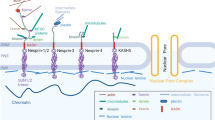
Mechanical forces generated by nuclear-cytoskeletal coupling through the linker of nucleoskeleton and cytoskeleton (LINC) complex, an evolutionarily conserved molecular bridge in the nuclear envelope (NE), are critical for the execution of wholesale nuclear positioning events in migrating and dividing cells, chromosome dynamics during meiosis, and mechanotransduction. LINC complexes consist of outer Klarsicht, ANC-1, and Syne homology (KASH) and inner Sad1, UNC-84 (SUN) nuclear membrane proteins. KASH proteins interact with the cytoskeleton in the cytoplasm and SUN proteins in the perinuclear space of the NE. In the nucleoplasm, SUN proteins interact with A-type nuclear lamins and chromatin-binding proteins. Recent structural insights into the KASH-SUN interaction have generated several questions regarding how LINC complex assembly and function might be regulated within the perinuclear space. Here we discuss potential LINC regulatory mechanisms and focus on the potential role of the ATPases associated with various cellular activities (AAA+) protein, torsinA, as a LINC complex regulator within the NE. We also examine how defects in LINC complex regulation by torsinA may contribute to the pathogenesis of the human neurological movement disorder, DYT1 dystonia.



Similar content being viewed by others
References
Albanese, A., et al. Phenomenology and classification of dystonia: a consensus update. Mov. Disord. 28:863–873, 2013.
Appenzeller-Herzog, C., and L. Ellgaard. The human PDI family: versatility packed into a single fold. Biochim. Biophys. Acta 1783:535–548, 2008.
Breakefield, X. O., C. Kamm, and P. I. Hanson. TorsinA: movement at many levels. Neuron 31:9–12, 2001.
Breakefield, X. O., et al. The pathophysiological basis of dystonias. Nat. Rev. Neurosci. 9:222–234, 2008.
Bressman, S. B., et al. Idiopathic dystonia among Ashkenazi Jews: evidence for autosomal dominant inheritance. Ann. Neurol. 26:612–620, 1989.
Brown, R. S., C. Zhao, A. R. Chase, J. Wang, and C. Schlieker. The mechanism of Torsin ATPase activation. Proc. Natl. Acad. Sci. USA 111:E4822–E4831, 2014.
Burdette, A. J., P. F. Churchill, G. A. Caldwell, and K. A. Caldwell. The early-onset torsion dystonia-associated protein, torsinA, displays molecular chaperone activity in vitro. Cell Stress Chaperones 15:605–617, 2010.
Burke, B. The nuclear envelope: filling in gaps. Nat. Cell Biol. 3:E273–E274, 2001.
Cain, N. E., and D. A. Starr. SUN proteins and nuclear envelope spacing. Nucleus 6(1):2–7, 2014.
Callan, A. C., S. Bunning, O. T. Jones, S. High, and E. Swanton. Biosynthesis of the dystonia-associated AAA + ATPase torsinA at the endoplasmic reticulum. Biochem. J. 401:607–612, 2007.
Chang, W., H. J. Worman, and G. G. Gundersen. Accessorizing and anchoring the LINC complex for multifunctionality. J. Cell Biol. 208:11–22, 2015.
Crisp, M., et al. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 172:41–53, 2006.
de Anda, F. C., et al. Centrosome localization determines neuronal polarity. Nature 436:704–708, 2005.
Dorboz, I., et al. Severe dystonia, cerebellar atrophy, and cardiomyopathy likely caused by a missense mutation in TOR1AIP1. Orphanet J. Rare Dis. 9:174, 2014.
Duong, N. T., et al. Nesprins: tissue-specific expression of epsilon and other short isoforms. PLoS One 9:e94380, 2014.
Epidemiological, S. O. D. I. E. E. S. D. E. C. G. A prevalence study of primary dystonia in eight European countries. J. Neurol. 247:787–792, 2000.
Fahn, S. Concept and classification of dystonia. Adv. Neurol. 50:1–8, 1988.
Gerace, L. TorsinA and torsion dystonia: unraveling the architecture of the nuclear envelope. Proc. Natl. Acad. Sci. USA 101:8839–8840, 2004.
Giles, L. M., J. Chen, L. Li, and L. S. Chin. Dystonia-associated mutations cause premature degradation of torsinA protein and cell-type-specific mislocalization to the nuclear envelope. Hum. Mol. Genet. 17:2712–2722, 2008.
Goodchild, R. E., and W. T. Dauer. Mislocalization to the nuclear envelope: an effect of the dystonia-causing torsinA mutation. Proc. Natl. Acad. Sci. USA 101:847–852, 2004.
Goodchild, R. E., and W. T. Dauer. The AAA + protein torsinA interacts with a conserved domain present in LAP1 and a novel ER protein. J. Cell Biol. 168:855–862, 2005.
Goodchild, R. E., C. E. Kim, and W. T. Dauer. Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron 48:923–932, 2005.
Goodchild, R. E., et al. Access of torsinA to the inner nuclear membrane is activity dependent and regulated in the endoplasmic reticulum. J. Cell Sci. 128:2854–2865, 2015.
Gordon, K. L., and P. Gonzalez-Alegre. Consequences of the DYT1 mutation on torsinA oligomerization and degradation. Neuroscience 157:588–595, 2008.
Hanson, P. I., and S. W. Whiteheart. AAA + proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6:519–529, 2005.
Hewett, J. W., et al. TorsinB–perinuclear location and association with torsinA. J. Neurochem. 89:1186–1194, 2004.
Hiraoka, Y., and A. F. Dernburg. The SUN rises on meiotic chromosome dynamics. Dev. Cell 17:598–605, 2009.
Iyer, L. M., D. D. Leipe, E. V. Koonin, and L. Aravind. Evolutionary history and higher order classification of AAA + ATPases. J. Struct. Biol. 146:11–31, 2004.
Jahed, Z., H. Shams, and M. R. Mofrad. A disulfide bond is required for the transmission of forces through SUN-KASH complexes. Biophys. J. 109:501–509, 2015.
Jokhi, V., et al. Torsin mediates primary envelopment of large ribonucleoprotein granules at the nuclear envelope. Cell Rep. 3:988–995, 2013.
Jungwirth, M., M. L. Dear, P. Brown, K. Holbrook, and R. Goodchild. Relative tissue expression of homologous torsinB correlates with the neuronal specific importance of DYT1 dystonia-associated torsinA. Hum. Mol. Genet. 19:888–900, 2010.
Kim, C. E., A. Perez, G. Perkins, M. H. Ellisman, and W. T. Dauer. A molecular mechanism underlying the neural-specific defect in torsinA mutant mice. Proc. Natl. Acad. Sci. USA 107:9861–9866, 2010.
Konakova, M., and S. M. Pulst. Dystonia-associated forms of torsinA are deficient in ATPase activity. J. Mol. Neurosci. 25:105–117, 2005.
Kustedjo, K., M. H. Bracey, and B. F. Cravatt. Torsin A and its torsion dystonia-associated mutant forms are lumenal glycoproteins that exhibit distinct subcellular localizations. J. Biol. Chem. 275:27933–27939, 2000.
Kustedjo, K., S. Deechongkit, J. W. Kelly, and B. F. Cravatt. Recombinant expression, purification, and comparative characterization of torsinA and its torsion dystonia-associated variant Delta E-torsinA. Biochemistry 42:15333–15341, 2003.
Li, R., and G. G. Gundersen. Beyond polymer polarity: how the cytoskeleton builds a polarized cell. Nat. Rev. Mol. Cell Biol. 9:860–873, 2008.
Liang, C. C., L. M. Tanabe, S. Jou, F. Chi, and W. T. Dauer. TorsinA hypofunction causes abnormal twisting movements and sensorimotor circuit neurodegeneration. J. Clin. Investig. 124:3080–3092, 2014.
Luxton, G. W., E. R. Gomes, E. S. Folker, E. Vintinner, and G. G. Gundersen. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science 329:956–959, 2010.
Luxton, G. W., E. R. Gomes, E. S. Folker, H. J. Worman, and G. G. Gundersen. TAN lines: a novel nuclear envelope structure involved in nuclear positioning. Nucleus 2:173–181, 2011.
Luxton, G. W., and G. G. Gundersen. Orientation and function of the nuclear-centrosomal axis during cell migration. Curr. Opin. Cell Biol. 23:579–588, 2011.
Luxton, G. W., and D. A. Starr. KASHing up with the nucleus: novel functional roles of KASH proteins at the cytoplasmic surface of the nucleus. Curr. Opin. Cell Biol. 28:69–75, 2014.
Maric, M., et al. A functional role for TorsinA in herpes simplex virus 1 nuclear egress. J. Virol. 85:9667–9679, 2011.
McCarthy, D. M., V. Gioioso, X. Zhang, N. Sharma, and P. G. Bhide. Neurogenesis and neuronal migration in the forebrain of the TorsinA knockout mouse embryo. Dev. Neurosci. 34:366–378, 2012.
Meinke, P., T. D. Nguyen, and M. S. Wehnert. The LINC complex and human disease. Biochem. Soc. Trans. 39:1693–1697, 2011.
Meinke, P., and E. C. Schirmer. LINC’ing form and function at the nuclear envelope. FEBS Lett. 589(19):2514–2521, 2015.
Morris, G. E., and K. N. Randles. Nesprin isoforms: are they inside or outside the nucleus. Biochem. Soc. Trans. 38:278–280, 2010.
Nagy, M., H. C. Wu, Z. Liu, S. Kedzierska-Mieszkowska, and M. Zolkiewski. Walker-A threonine couples nucleotide occupancy with the chaperone activity of the AAA + ATPase ClpB. Protein Sci. 18:287–293, 2009.
Naismith, T. V., S. Dalal, and P. I. Hanson. Interaction of torsinA with its major binding partners is impaired by the dystonia-associated DeltaGAG deletion. J. Biol. Chem. 284:27866–27874, 2009.
Naismith, T. V., J. E. Heuser, X. O. Breakefield, and P. I. Hanson. TorsinA in the nuclear envelope. Proc. Natl. Acad. Sci. USA 101:7612–7617, 2004.
Nery, F. C., et al. TorsinA binds the KASH domain of nesprins and participates in linkage between nuclear envelope and cytoskeleton. J. Cell Sci. 121:3476–3486, 2008.
Nery, F. C., et al. TorsinA participates in endoplasmic reticulum-associated degradation. Nat. Commun. 2:393, 2011.
Nery, F. C., et al. Microfluidic platform to evaluate migration of cells from patients with DYT1 dystonia. J. Neurosci. Methods 232:181–188, 2014.
Neumann, S., and A. A. Noegel. Nesprins in cell stability and migration. Adv. Exp. Med. Biol. 773:491–504, 2014.
Ostlund, C., et al. Dynamics and molecular interactions of linker of nucleoskeleton and cytoskeleton (LINC) complex proteins. J. Cell Sci. 122:4099–4108, 2009.
Ozelius, L. J., and S. B. Bressman. Genetic and clinical features of primary torsion dystonia. Neurobiol. Dis. 42:127–135, 2011.
Ozelius, L. J., et al. The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat. Genet. 17:40–48, 1997.
Ozelius, L. J., et al. The TOR1A (DYT1) gene family and its role in early onset torsion dystonia. Genomics 62:377–384, 1999.
Packard, M., et al. Nucleus to synapse nesprin1 railroad tracks direct synapse maturation through RNA localization. Neuron 86:1015–1028, 2015.
Pham, P., K. P. Frei, W. Woo, and D. D. Truong. Molecular defects of the dystonia-causing torsinA mutation. Neuroreport 17:1725–1728, 2006.
Rose, A. E., C. Zhao, E. M. Turner, A. M. Steyer, and C. Schlieker. Arresting a Torsin ATPase reshapes the endoplasmic reticulum. J. Biol. Chem. 289:552–564, 2014.
Scheffzek, K., M. R. Ahmadian, and A. Wittinghofer. GTPase-activating proteins: helping hands to complement an active site. Trends Biochem. Sci. 23:257–262, 1998.
Shin, J. Y., et al. Lamina-associated polypeptide-1 interacts with the muscular dystrophy protein emerin and is essential for skeletal muscle maintenance. Dev. Cell 26:591–603, 2013.
Sosa, B. A., A. Rothballer, U. Kutay, and T. U. Schwartz. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell 149:1035–1047, 2012.
Sosa, B. A., et al. How lamina-associated polypeptide 1 (LAP1) activates Torsin. Elife 3:e03239, 2014.
Speese, S. D., et al. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell 149:832–846, 2012.
Starr, D. A., and M. Han. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science 298:406–409, 2002.
Stewart-Hutchinson, P. J., C. M. Hale, D. Wirtz, and D. Hodzic. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp. Cell Res. 314:1892–1905, 2008.
Swartz, R. K., E. C. Rodriguez, and M. C. King. A role for nuclear envelope-bridging complexes in homology-directed repair. Mol. Biol. Cell 25:2461–2471, 2014.
Torres, G. E., A. L. Sweeney, J. M. Beaulieu, P. Shashidharan, and M. G. Caron. Effect of torsinA on membrane proteins reveals a loss of function and a dominant-negative phenotype of the dystonia-associated DeltaE-torsinA mutant. Proc. Natl. Acad. Sci. USA 101:15650–15655, 2004.
Turner, E. M., R. S. Brown, E. Laudermilch, P. L. Tsai, and C. Schlieker. The Torsin activator LULL1 is required for efficient growth of HSV-1. J. Virol. 89(16):8444–8452, 2015.
Ulug, A. M., et al. Cerebellothalamocortical pathway abnormalities in torsinA DYT1 knock-in mice. Proc. Natl. Acad. Sci. USA 108:6638–6643, 2011.
Vale, R. D. AAA proteins. Lords of the ring. J. Cell Biol. 150:F13–F19, 2000.
Van der Heyden, A. B., T. V. Naismith, E. L. Snapp, and P. I. Hanson. Static retention of the lumenal monotopic membrane protein torsinA in the endoplasmic reticulum. EMBO J. 30:3217–3231, 2011.
Van der Heyden, A. B., T. V. Naismith, E. L. Snapp, D. Hodzic, and P. I. Hanson. LULL1 retargets TorsinA to the nuclear envelope revealing an activity that is impaired by the DYT1 dystonia mutation. Mol. Biol. Cell 20:2661–2672, 2009.
VanGompel, M. J., K. C. Nguyen, D. H. Hall, W. T. Dauer, and L. S. A. Rose. Novel function for the C. elegans torsin OOC-5 in nucleoporin localization and nuclear import. Mol. Biol. Cell 26(9):1752–1763, 2015.
Wang, W., et al. Structural insights into SUN-KASH complexes across the nuclear envelope. Cell Res. 22:1440–1452, 2012.
Weibezahn, J., C. Schlieker, B. Bukau, and A. Mogk. Characterization of a trap mutant of the AAA + chaperone ClpB. J. Biol. Chem. 278:32608–32617, 2003.
Worman, H. J., and W. T. Dauer. The nuclear envelope: an intriguing focal point for neurogenetic disease. Neurotherapeutics 11:764–772, 2014.
Zhang, X., et al. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron 64:173–187, 2009.
Zhao, C., R. S. Brown, A. R. Chase, M. R. Eisele, and C. Schlieker. Regulation of torsin ATPases by LAP1 and LULL1. Proc. Natl. Acad. Sci. USA 110:E1545–E1554, 2013.
Zhou, Z., et al. Structure of Sad1-UNC84 homology (SUN) domain defines features of molecular bridge in nuclear envelope. J. Biol. Chem. 287:5317–5326, 2012.
Zhu, L., L. Millen, J. L. Mendoza, and P. J. Thomas. A unique redox-sensing sensor II motif in TorsinA plays a critical role in nucleotide and partner binding. J. Biol. Chem. 285:37271–37280, 2010.
Zhu, L., J. O. Wrabl, A. P. Hayashi, L. S. Rose, and P. J. Thomas. The torsin-family AAA + protein OOC-5 contains a critical disulfide adjacent to Sensor-II that couples redox state to nucleotide binding. Mol. Biol. Cell 19:3599–3612, 2008.
Acknowledgments
We thank Drs. Melissa Gardner, David Greenstein, Joachim Mueller, and Meg Titus for helpful discussions. The authors apologize to those whose work we were unable to cite and cover in proper depth due to the limitations of length for this review. Studies in the Luxton Lab are supported by start up funding from the University of Minnesota, a P30 Pilot and Feasibility Grant from the Paul and Sheila Wellstone Muscular Dystrophy Center, and funding from the NIH (R21 NS095109-01, 1R41DA037622, and AR57220). C.A.S. is supported by an NIH training grant (NIH 5T32AR007612-14).
Conflict of interest
Cosmo A. Saunders and G.W. Gant Luxton have no conflicts on interest.
Ethical Standards
The authors for this article carried out neither animal nor human studies for this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Kris Noel Dahl oversaw the review of this article.
Rights and permissions
About this article
Cite this article
Saunders, C.A., Luxton, G.W.G. LINCing Defective Nuclear-Cytoskeletal Coupling and DYT1 Dystonia. Cel. Mol. Bioeng. 9, 207–216 (2016). https://doi.org/10.1007/s12195-016-0432-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12195-016-0432-0