Abstract
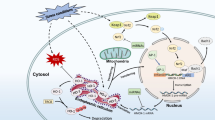
The hematopoietic transcription factor GATA1 induces heme accumulation during erythropoiesis by directly activating genes mediating heme biosynthesis. In addition to its canonical functions as a hemoglobin prosthetic group and enzyme cofactor, heme regulates gene expression in erythroid cells both transcriptionally and post-transcriptionally. Heme binding to the transcriptional repressor BACH1 triggers its proteolytic degradation. In heme-deficient cells, BACH1 accumulates and represses transcription of target genes, including α- and β-like globin genes, preventing the accumulation of cytotoxic free globin chains. A recently described BACH1-independent mechanism of heme-dependent transcriptional regulation is associated with a DNA motif termed heme-regulated motif (HERM), which resides at the majority of loci harboring heme-regulated chromatin accessibility sites. Progress on these problems has led to a paradigm in which cell type-specific transcriptional mechanisms determine the expression of enzymes mediating the synthesis of small molecules, which generate feedback loops, converging upon the transcription factor itself and the genome. This marriage between transcription factors and the small molecules that they control is predicted to be a canonical attribute of regulatory networks governing cell state transitions such as differentiation in the hematopoietic system and more broadly.




Similar content being viewed by others
References
Dutt S, Hamza I, Bartnikas TB. Molecular mechanisms of iron and heme metabolism. Annu Rev Nutr. 2022. https://doi.org/10.1146/annurev-nutr-062320-112625.
Dailey HA, Meissner PN. Erythroid heme biosynthesis and its disorders. Cold Spring Harb Perspect Med. 2013;3(4): a011676.
Yuan X, Fleming MD, Hamza I. Heme transport and erythropoiesis. Curr Opin Chem Biol. 2013;17(2):204–11.
Ajioka RS, Phillips JD, Kushner JP. Biosynthesis of heme in mammals. Biochim Biophys Acta. 2006;1763(7):723–36.
Bishop DF, Henderson AS, Astrin KH. Human delta-aminolevulinate synthase: assignment of the housekeeping gene to 3p21 and the erythroid-specific gene to the X chromosome. Genomics. 1990;7(2):207–14.
Tanimura N, Miller E, Igarashi K, Yang D, Burstyn JN, Dewey CN, et al. Mechanism governing heme synthesis reveals a GATA factor/heme circuit that controls differentiation. EMBO Rep. 2016;17(2):249–65.
Campagna DR, de Bie CI, Schmitz-Abe K, Sweeney M, Sendamarai AK, Schmidt PJ, et al. X-linked sideroblastic anemia due to ALAS2 intron 1 enhancer element GATA-binding site mutations. Am J Hematol. 2014;89(3):315–9.
Kaneko K, Furuyama K, Fujiwara T, Kobayashi R, Ishida H, Harigae H, et al. Identification of a novel erythroid-specific enhancer for the ALAS2 gene and its loss-of-function mutation which is associated with congenital sideroblastic anemia. Haematologica. 2014;99(2):252–61.
Zhang Y, Zhang J, An W, Wan Y, Ma S, Yin J, et al. Intron 1 GATA site enhances ALAS2 expression indispensably during erythroid differentiation. Nucleic Acids Res. 2017;45(2):657–71.
Kruger A, Keppel M, Sharma V, Frunzke J. The diversity of heme sensor systems—heme-responsive transcriptional regulation mediated by transient heme protein interactions. FEMS Microbiol Rev. 2022;46(3): fuac002.
Shimizu T, Lengalova A, Martinek V, Martinkova M. Heme: emergent roles of heme in signal transduction, functional regulation and as catalytic centres. Chem Soc Rev. 2019;48(24):5624–57.
Chen JJ. Regulation of protein synthesis by the heme-regulated eIF2alpha kinase: relevance to anemias. Blood. 2007;109(7):2693–9.
Chen JJ, Zhang S. Translational control by heme-regulated elF2alpha kinase during erythropoiesis. Curr Opin Hematol. 2022;29(3):103–11.
Han AP, Yu C, Lu L, Fujiwara Y, Browne C, Chin G, et al. Heme-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J. 2001;20(23):6909–18.
Tahara T, Sun J, Igarashi K, Taketani S. Heme-dependent up-regulation of the alpha-globin gene expression by transcriptional repressor Bach1 in erythroid cells. Biochem Biophys Res Commun. 2004;324(1):77–85.
Tahara T, Sun J, Nakanishi K, Yamamoto M, Mori H, Saito T, et al. Heme positively regulates the expression of beta-globin at the locus control region via the transcriptional factor Bach1 in erythroid cells. J Biol Chem. 2004;279(7):5480–7.
Oyake T, Itoh K, Motohashi H. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol. 1996;16:6083–95.
Ogawa K, Sun J, Taketani S, Nakajima O, Nishitani C, Sassa S, et al. Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J. 2001;20(11):2835–43.
Lathrop JT, Timko MP. Regulation by heme of mitochondrial protein transport through a conserved amino acid motif. Science. 1993;259(5094):522–5.
Munakata H, Sun JY, Yoshida K, Nakatani T, Honda E, Hayakawa S, et al. Role of the heme regulatory motif in the heme-mediated inhibition of mitochondrial import of 5-aminolevulinate synthase. J Biochem. 2004;136(2):233–8.
Igarashi J, Murase M, Iizuka A, Pichierri F, Martinkova M, Shimizu T. Elucidation of the heme binding site of heme-regulated eukaryotic initiation factor 2alpha kinase and the role of the regulatory motif in heme sensing by spectroscopic and catalytic studies of mutant proteins. J Biol Chem. 2008;283(27):18782–91.
Hira S, Tomita T, Matsui T, Igarashi K, Ikeda-Saito M. Bach1, a heme-dependent transcription factor, reveals presence of multiple heme binding sites with distinct coordination structure. IUBMB Life. 2007;59(8–9):542–51.
Segawa K, Igarashi K, Murayama K. The Cys-Pro motifs in the intrinsically disordered regions of the transcription factor BACH1 mediate distinct and overlapping functions upon heme binding. FEBS Lett. 2022. https://doi.org/10.1002/1873-3468.14338.
Sun J, Hoshino H, Takaku K, Nakajima O, Muto A, Suzuki H, et al. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 2002;21(19):5216–24.
Sun J, Brand M, Zenke Y, Tashiro S, Groudine M, Igarashi K. Heme regulates the dynamic exchange of Bach1 and NF-E2-related factors in the Maf transcription factor network. Proc Natl Acad Sci USA. 2004;101(6):1461–6.
Suzuki H, Tashiro S, Hira S, Sun J, Yamazaki C, Zenke Y, et al. Heme regulates gene expression by triggering Crm1-dependent nuclear export of Bach1. EMBO J. 2004;23(13):2544–53.
Suzuki H, Tashiro S, Sun J, Doi H, Satomi S, Igarashi K. Cadmium induces nuclear export of Bach1, a transcriptional repressor of heme oxygenase-1 gene. J Biol Chem. 2003;278(49):49246–53.
Zenke-Kawasaki Y, Dohi Y, Katoh Y, Ikura T, Ikura M, Asahara T, et al. Heme induces ubiquitination and degradation of the transcription factor Bach1. Mol Cell Biol. 2007;27(19):6962–71.
Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 2006;25(20):4877–87.
Boisson B, Laplantine E, Prando C, Giliani S, Israelsson E, Xu Z, et al. Immunodeficiency, autoinflammation and amylopectinosis in humans with inherited HOIL-1 and LUBAC deficiency. Nat Immunol. 2012;13(12):1178–86.
Zinngrebe J, Rieser E, Taraborrelli L, Peltzer N, Hartwig T, Ren H, et al. LUBAC deficiency perturbs TLR3 signaling to cause immunodeficiency and autoinflammation. J Exp Med. 2016;213(12):2671–89.
Tan MK, Lim HJ, Bennett EJ, Shi Y, Harper JW. Parallel SCF adaptor capture proteomics reveals a role for SCFFBXL17 in NRF2 activation via BACH1 repressor turnover. Mol Cell. 2013;52(1):9–24.
Lignitto L, LeBoeuf SE, Homer H, Jiang S, Askenazi M, Karakousi TR, et al. Nrf2 activation promotes lung cancer metastasis by inhibiting the degradation of Bach1. Cell. 2019;178(2):316-329 e18.
Mouse EC, Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, et al. An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol. 2012;13(8):418.
Warnatz HJ, Schmidt D, Manke T, Piccini I, Sultan M, Borodina T, et al. The BTB and CNC homology 1 (BACH1) target genes are involved in the oxidative stress response and in control of the cell cycle. J Biol Chem. 2011;286(26):23521–32.
Matsumoto M, Kondo K, Shiraki T, Brydun A, Funayama R, Nakayama K, et al. Genomewide approaches for BACH1 target genes in mouse embryonic fibroblasts showed BACH1-Pparg pathway in adipogenesis. Genes Cells. 2016;21(6):553–67.
Ebina-Shibuya R, Watanabe-Matsui M, Matsumoto M, Itoh-Nakadai A, Funayama R, Nakayama K, et al. The double knockout of Bach1 and Bach2 in mice reveals shared compensatory mechanisms in regulating alveolar macrophage function and lung surfactant homeostasis. J Biochem. 2016;160(6):333–44.
Igarashi K, Itoh-Nakadai A. Orchestration of B lymphoid cells and their inner myeloid by Bach. Curr Opin Immunol. 2016;39:136–42.
Watanabe-Matsui M, Matsumoto T, Matsui T, Ikeda-Saito M, Muto A, Murayama K, et al. Heme binds to an intrinsically disordered region of Bach2 and alters its conformation. Arch Biochem Biophys. 2015;565:25–31.
Watanabe-Matsui M, Muto A, Matsui T, Itoh-Nakadai A, Nakajima O, Murayama K, et al. Heme regulates B-cell differentiation, antibody class switch, and heme oxygenase-1 expression in B cells as a ligand of Bach2. Blood. 2011;117(20):5438–48.
Kato H, Itoh-Nakadai A, Matsumoto M, Ishii Y, Watanabe-Matsui M, Ikeda M, et al. Infection perturbs Bach2- and Bach1-dependent erythroid lineage “choice” to cause anemia. Nat Immunol. 2018;19(10):1059–70.
Weiss MJ, Yu C, Orkin SH. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol. 1997;17(3):1642–51.
Gregory T, Yu C, Ma A, Orkin SH, Blobel GA, Weiss MJ. GATA-1 and erythropoietin cooperate to promoter erythroid cell survival by regulating bcl-xl expression. Blood. 1999;94(1):87–96.
Grass JA, Boyer ME, Pal S, Wu J, Weiss MJ, Bresnick EH. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc Natl Acad Sci USA. 2003;100:8811–6.
Welch JJ, Watts JA, Vakoc CR, Yao Y, Wang H, Hardison RC, et al. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104:3136–47.
Tanimura N, Liao R, Wilson GM, Dent MR, Cao M, Burstyn JN, et al. GATA/heme multi-omics reveals a trace metal-dependent cellular differentiation mechanism. Dev Cell. 2018;46(5):581-594 e4.
Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10(12):1213–8.
Liao R, Zheng Y, Liu X, Zhang Y, Seim G, Tanimura N, et al. Discovering how heme controls genome function through heme-omics. Cell Rep. 2020;31(13): 107832.
Noguchi CT, Rodgers GP, Serjeant G, Schechter AN. Predicting therapeutic efficacy for sickle cell anemia. Prog Clin Biol Res. 1987;251:497–505.
Baronciani L, Beutler E. Analysis of pyruvate kinase-deficiency mutations that produce nonspherocytic hemolytic anemia. Proc Natl Acad Sci USA. 1993;90(9):4324–7.
Frietze S, O’Geen H, Blahnik KR, Jin VX, Farnham PJ. ZNF274 recruits the histone methyltransferase SETDB1 to the 3′ ends of ZNF genes. PLoS One. 2010;5(12): e15082.
Quigley JG, Yang Z, Worthington MT, Phillips JD, Sabo KM, Sabath DE, et al. Identification of a human heme exporter that is essential for erythropoiesis. Cell. 2004;118(6):757–66.
Doty RT, Phelps SR, Shadle C, Sanchez-Bonilla M, Keel SB, Abkowitz JL. Coordinate expression of heme and globin is essential for effective erythropoiesis. J Clin Investig. 2015;125(12):4681–91.
Keel SB, Doty RT, Yang Z, Quigley JG, Chen J, Knoblaugh S, et al. A heme export protein is required for red blood cell differentiation and iron homeostasis. Science. 2008;319(5864):825–8.
Doty RT, Yan X, Lausted C, Munday AD, Yang Z, Yi D, et al. Single-cell analyses demonstrate that a heme-GATA1 feedback loop regulates red cell differentiation. Blood. 2019;133(5):457–69.
Suragani RN, Zachariah RS, Velazquez JG, Liu S, Sun CW, Townes TM, et al. Heme-regulated eIF2alpha kinase activated Atf4 signaling pathway in oxidative stress and erythropoiesis. Blood. 2012;119(22):5276–84.
Zhang S, Macias-Garcia A, Velazquez J, Paltrinieri E, Kaufman RJ, Chen JJ. HRI coordinates translation by eIF2alphaP and mTORC1 to mitigate ineffective erythropoiesis in mice during iron deficiency. Blood. 2018;131(4):450–61.
Wortel IMN, van der Meer LT, Kilberg MS, van Leeuwen FN. Surviving stress: modulation of ATF4-mediated stress responses in normal and malignant cells. Trends Endocrinol Metab. 2017;28(11):794–806.
Zhang S, Macias-Garcia A, Ulirsch JC, Velazquez J, Butty VL, Levine SS, et al. HRI coordinates translation necessary for protein homeostasis and mitochondrial function in erythropoiesis. Elife. 2019;8: e46976.
Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol. 2013;15(5):481–90.
Pathak SS, Liu D, Li T, de Zavalia N, Zhu L, Li J, et al. The eIF2alpha kinase GCN2 modulates period and rhythmicity of the circadian clock by translational control of Atf4. Neuron. 2019;104(4):724-35 e6.
Zwifelhofer NM, Cai X, Liao R, Mao B, Conn DJ, Mehta C, et al. GATA factor-regulated solute carrier ensemble reveals a nucleoside transporter-dependent differentiation mechanism. PLoS Genet. 2020;16(12): e1009286.
Acknowledgements
This work is supported by NIH grant R01DK50107 and Carbone Cancer Center P30CA014520.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
About this article
Cite this article
Liao, R., Bresnick, E.H. Heme as a differentiation-regulatory transcriptional cofactor. Int J Hematol 116, 174–181 (2022). https://doi.org/10.1007/s12185-022-03404-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12185-022-03404-x